Review: Synthetic algal genomes
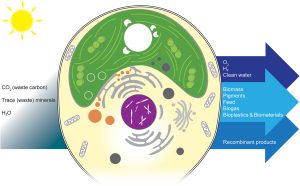 As photosynthetic organisms, algae can harness solar energy to do useful things, from environmental cleanup to producing fuels and other beneficial molecules. This review by Goold et al. provides an overview of how algae can be valuable platforms for synthetic biology and metabolic engineering through the incorporation of synthetic genomes. The authors build on the work of the Synthetic Yeast Genome Project (Sc2.0), which endeavors to redesign the yeast genome to make it more concise and easier to manipulate (for example, by introducing recombination sites and adding “synthetic supernumerary neochromosomes”); basically, if you want to use a cell as a factory, start by tidying up the junk and making it into an environment designed for efficiency. The best system for a synthetic algal genome project isn’t quite as clearcut as yeast. The authors first look at the potential of Chlamydomonas reinhardtii, the algal superstar. However good it is as a model system, it isn’t optimal for this approach due to its large genome, so they then discuss other algal species as candidates for synthetic genomes. Beneficial traits include rapid growth, both diploid and haploid states with the possibility of sexual reproduction, interesting metabolic pathways, and the ability to grow heterotrophically, particularly when photosynthetic genes are being manipulated. Given the great diversity of algae, there are several good candidates and it seems likely that as this technology develops many will be employed. This is a very thought-provoking article that provides a peek into the future of algal biotechnology. (Summary by Mary Williams @PlantTeaching) Cell Genomics 10.1016/j.xgen.2024.100505
As photosynthetic organisms, algae can harness solar energy to do useful things, from environmental cleanup to producing fuels and other beneficial molecules. This review by Goold et al. provides an overview of how algae can be valuable platforms for synthetic biology and metabolic engineering through the incorporation of synthetic genomes. The authors build on the work of the Synthetic Yeast Genome Project (Sc2.0), which endeavors to redesign the yeast genome to make it more concise and easier to manipulate (for example, by introducing recombination sites and adding “synthetic supernumerary neochromosomes”); basically, if you want to use a cell as a factory, start by tidying up the junk and making it into an environment designed for efficiency. The best system for a synthetic algal genome project isn’t quite as clearcut as yeast. The authors first look at the potential of Chlamydomonas reinhardtii, the algal superstar. However good it is as a model system, it isn’t optimal for this approach due to its large genome, so they then discuss other algal species as candidates for synthetic genomes. Beneficial traits include rapid growth, both diploid and haploid states with the possibility of sexual reproduction, interesting metabolic pathways, and the ability to grow heterotrophically, particularly when photosynthetic genes are being manipulated. Given the great diversity of algae, there are several good candidates and it seems likely that as this technology develops many will be employed. This is a very thought-provoking article that provides a peek into the future of algal biotechnology. (Summary by Mary Williams @PlantTeaching) Cell Genomics 10.1016/j.xgen.2024.100505