Review Single-cell genomics and epigenomics: Technologies and applications in plants ($) (Trends Plant Sci)
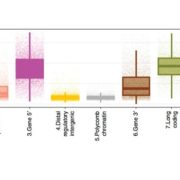
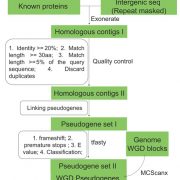
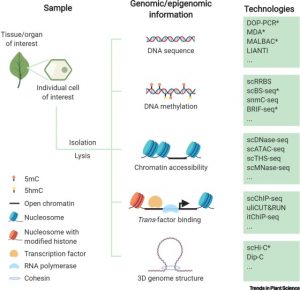 Plants (embryophytes) are by definition multicellular, but we seek to understand them as the sum of the activities of individual cells. Much of this knowledge rests on information obtained through grinding up tissues made up from several cell types. This review by Luo et al. describes methods for plant single-cell genomics and epigenomics studies. All involve first isolating single cells, followed by whole-genome amplification to obtain sufficient material for analysis. As many of the techniques were developed for use with easy-to-disperse animal cells, the additional steps required to dissociate plant cells adds potential for artifacts. At the genomic sequence level, single-cell methods can point to single-nucleotide as well as copy-number variations. A suite of epigenomic analysis are also possible, including analysis of DNA methylation and even DNA-histone interactions, though as yet it isn’t possible to survey transcription factor binding within single cells. The authors describe some applications of these technologies, and also point to the promise of complete -omics datasets for every cell type. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci. 10.1016/j.tplants.2020.04.016
Plants (embryophytes) are by definition multicellular, but we seek to understand them as the sum of the activities of individual cells. Much of this knowledge rests on information obtained through grinding up tissues made up from several cell types. This review by Luo et al. describes methods for plant single-cell genomics and epigenomics studies. All involve first isolating single cells, followed by whole-genome amplification to obtain sufficient material for analysis. As many of the techniques were developed for use with easy-to-disperse animal cells, the additional steps required to dissociate plant cells adds potential for artifacts. At the genomic sequence level, single-cell methods can point to single-nucleotide as well as copy-number variations. A suite of epigenomic analysis are also possible, including analysis of DNA methylation and even DNA-histone interactions, though as yet it isn’t possible to survey transcription factor binding within single cells. The authors describe some applications of these technologies, and also point to the promise of complete -omics datasets for every cell type. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci. 10.1016/j.tplants.2020.04.016