Review: Proximity labeling in plants
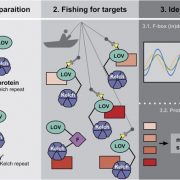
Genetic studies can suggest that two proteins function in the same pathway, but how can we figure out if they share the same space? In this review, Xu et al. provide an overview of proximity labeling, a method to identify proteins that co-localize in space. Proximity labeling uses a biotin ligase which is incubated with biotin to produce short-lived diffusible biotin adenylate intermediates. These tags are transferred to proteins within a very narrow (10nm) radius, and then the labeled proteins can be captured by streptavidin beads and identified through mass spectrometry; it’s a bit like throwing a tiny net into the cell and analyzing what is caught associating with what. The ligase can be fused to a protein of interest (bait) to search for its interactors, or targeted to an organelle, or expressed from a cell-specific promoter to query specific proteomes. These approaches have been enhanced by directed engineering of the biotin kinase enzyme to make it work faster, resulting in enzymes such as TurboID that can carry out proximity labeling in as little as ten minutes. This review focuses on the application of these methods in plants. (Summary by Mary Williams @PlantTeaching) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-070522-052132