Review: Integrative framework for successful deployment of microbiomes in agriculture
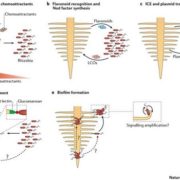
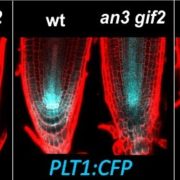
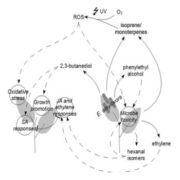
 This review by Berruto and Demirer calls for an interdisciplinary approach at the interface of experimental and computational frameworks to address the long-standing challenge of field application of beneficial soil microbiomes. This challenge persists due to our limited understanding of the complex principles governing plant-microbe and microbe-microbe interactions. The authors provide a comprehensive overview of both plant and microbial factors that determine this plant-microbe detente and proposed strategies to design microbial consortia for predictable plant phenotypes. Combining genome-wide association studies (GWAS) of host plants with metagenome-wide association studies (MWAS) could reveal target loci for plant breeding and improved plant-microbe associations. The authors propose a shift in our approach to integrating such datasets by considering feature interactions of both host and microbiome, especially through use of machine learning (ML). Conversely, the complexity of microbial interactions (pairwise or higher order) and their emergent properties could be mapped by developing ML based predictive frameworks incorporating data from co-inoculation experiments (putting two strains together and assessing plant phenotypes) with genome scale metabolic models (GSMMs). Finally, these strategies could be supplemented with emerging engineering techniques for both the host plant and its microbiome to precisely manipulate plant phenotypes. (Summary by Arijit Mukherjee @ArijitM61745830) Trends Microbiol. https://doi.org/10.1016/j.tim.2024.02.003
This review by Berruto and Demirer calls for an interdisciplinary approach at the interface of experimental and computational frameworks to address the long-standing challenge of field application of beneficial soil microbiomes. This challenge persists due to our limited understanding of the complex principles governing plant-microbe and microbe-microbe interactions. The authors provide a comprehensive overview of both plant and microbial factors that determine this plant-microbe detente and proposed strategies to design microbial consortia for predictable plant phenotypes. Combining genome-wide association studies (GWAS) of host plants with metagenome-wide association studies (MWAS) could reveal target loci for plant breeding and improved plant-microbe associations. The authors propose a shift in our approach to integrating such datasets by considering feature interactions of both host and microbiome, especially through use of machine learning (ML). Conversely, the complexity of microbial interactions (pairwise or higher order) and their emergent properties could be mapped by developing ML based predictive frameworks incorporating data from co-inoculation experiments (putting two strains together and assessing plant phenotypes) with genome scale metabolic models (GSMMs). Finally, these strategies could be supplemented with emerging engineering techniques for both the host plant and its microbiome to precisely manipulate plant phenotypes. (Summary by Arijit Mukherjee @ArijitM61745830) Trends Microbiol. https://doi.org/10.1016/j.tim.2024.02.003