Review. Endosperm variability: From endoreduplication within a seed to higher ploidy across species, and its competence ($) (Seed Sci. Res.)
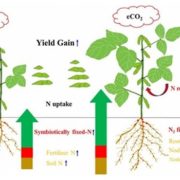
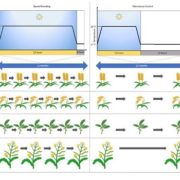
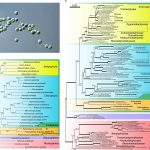
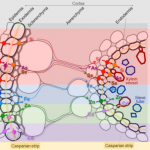
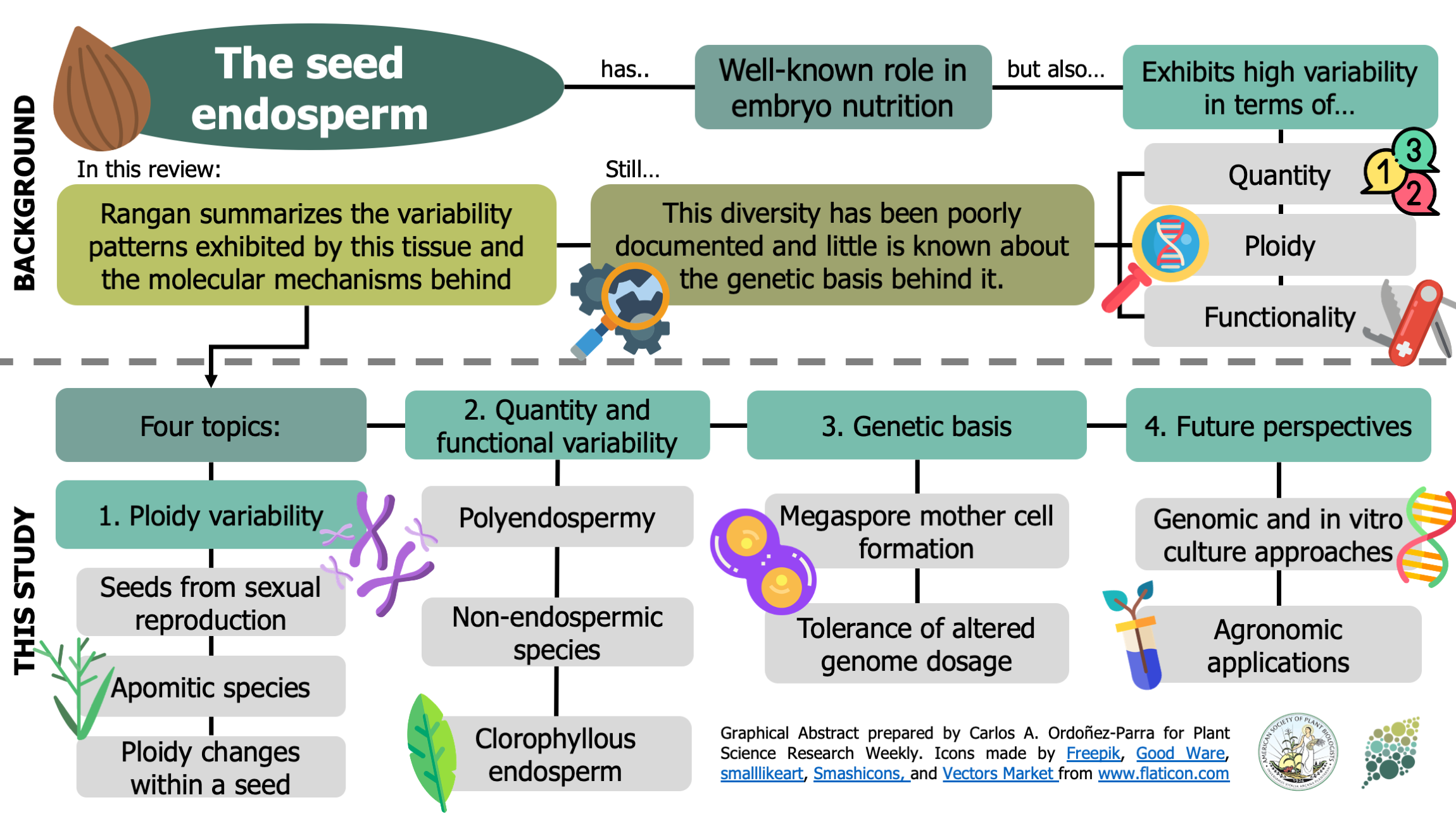 In addition to its well-known nutritive function, the endosperm of seeds has a great deal of morphological, genomic and functional diversity. In this review, Rangan summarizes endosperm variability and the molecular mechanisms behind it. Although classically described as being triploid (2n from the mother and n from the father), in reality endosperm ploidy is hugely variable across species, spanning from 2n-15n. Furthermore, paternal and maternal contributions differ between seeds that originate from sexual reproduction from those arising from the different forms of apomixis. Additionally, endoreduplication within the endosperm can increase its variability within a seed. Current information about the genetic basis behind this variability is discussed, emphasizing the potential roles of genes that regulate the cell cycle in endosperm development. The author observes that our knowledge about endosperm development is highly limited and largely based on Arabidopsis thaliana. The review concludes by outlining a “roadmap for understanding the endosperm genome variability,” highlighting the genomics and in vitro tissue culture approaches that can increase our understanding of the origin of this tissue diversity, and the potential agricultural applications that can arise in the process. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Seed Sci. Res. 10.1017/S0960258520000148
In addition to its well-known nutritive function, the endosperm of seeds has a great deal of morphological, genomic and functional diversity. In this review, Rangan summarizes endosperm variability and the molecular mechanisms behind it. Although classically described as being triploid (2n from the mother and n from the father), in reality endosperm ploidy is hugely variable across species, spanning from 2n-15n. Furthermore, paternal and maternal contributions differ between seeds that originate from sexual reproduction from those arising from the different forms of apomixis. Additionally, endoreduplication within the endosperm can increase its variability within a seed. Current information about the genetic basis behind this variability is discussed, emphasizing the potential roles of genes that regulate the cell cycle in endosperm development. The author observes that our knowledge about endosperm development is highly limited and largely based on Arabidopsis thaliana. The review concludes by outlining a “roadmap for understanding the endosperm genome variability,” highlighting the genomics and in vitro tissue culture approaches that can increase our understanding of the origin of this tissue diversity, and the potential agricultural applications that can arise in the process. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Seed Sci. Res. 10.1017/S0960258520000148