Review: Defining and rewiring of gene regulatory networks for plant improvement
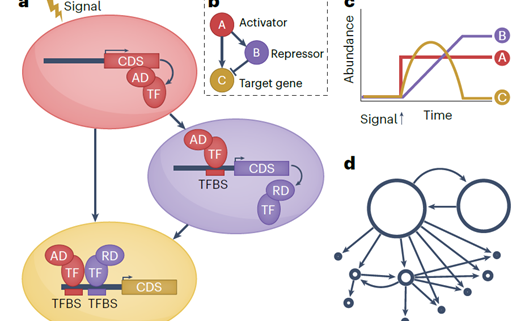
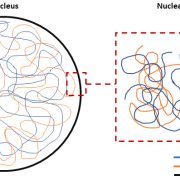
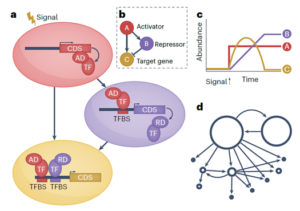 Much of the focus of functional genomics studies in plants is to improve yield, disease resistance, abiotic stress tolerance and nutritional quality. Many desirable traits are controlled by sets of genes that are coordinated in a complex network, called a gene regulatory network (GRN). A transcription factor (TF) binds to the promoter region of one or more genes and promotes or represses gene expression. In this review, Borowsky and Bailey-Serres discuss how defining the GRNs underlying desired traits can be used to fine-tune and further adjust them to meet the demands of climate change and food supply. Next-generation sequencing technologies coupled with omics studies made it possible to explore the gene circuitry and interacting proteins controlling favorable traits. The functions of most major genes are conserved among different species with much diversity attributed to TFs and cis-regulatory elements, which drive or alter the expression of target genes. For example, the function of the root TF SHORTROOT is conserved, but its redeployment results in the addition of more cortical cells in maize compared to Arabidopsis. Similarly, the function of the bundle sheath TF DNA BINDING WITH ONE FINGER is conserved in photosynthesis, however, its downstream deployment regulates C3 genes in rice and C4 genes in sorghum. The authors present a set of examples of how synthetic biology and genome editing can be used to engineer desirable phenotypes in plants through rewiring gene circuitry. (Summary by Asif Ali @pbgasifkalas) Nat. Genet. 10.1038/s41588-024-01806-7
Much of the focus of functional genomics studies in plants is to improve yield, disease resistance, abiotic stress tolerance and nutritional quality. Many desirable traits are controlled by sets of genes that are coordinated in a complex network, called a gene regulatory network (GRN). A transcription factor (TF) binds to the promoter region of one or more genes and promotes or represses gene expression. In this review, Borowsky and Bailey-Serres discuss how defining the GRNs underlying desired traits can be used to fine-tune and further adjust them to meet the demands of climate change and food supply. Next-generation sequencing technologies coupled with omics studies made it possible to explore the gene circuitry and interacting proteins controlling favorable traits. The functions of most major genes are conserved among different species with much diversity attributed to TFs and cis-regulatory elements, which drive or alter the expression of target genes. For example, the function of the root TF SHORTROOT is conserved, but its redeployment results in the addition of more cortical cells in maize compared to Arabidopsis. Similarly, the function of the bundle sheath TF DNA BINDING WITH ONE FINGER is conserved in photosynthesis, however, its downstream deployment regulates C3 genes in rice and C4 genes in sorghum. The authors present a set of examples of how synthetic biology and genome editing can be used to engineer desirable phenotypes in plants through rewiring gene circuitry. (Summary by Asif Ali @pbgasifkalas) Nat. Genet. 10.1038/s41588-024-01806-7