Reprogramming of root cells during nitrogen-fixing symbiosis involves dynamic polysome association of coding and noncoding RNAs ($) (Plant Cell)
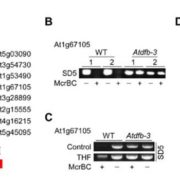
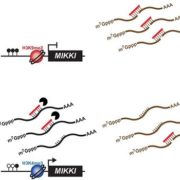
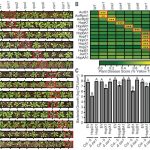
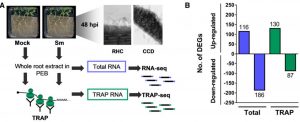 The symbiotic relationship between Rhizobium bacteria and leguminous plants like Medicago results in the development of secondary root organs called nodules. The bacteria housed in the nodule infection zone assimilate atmospheric nitrogen for plant growth. In this paper, Traubenik et al. used (RNA-seq) and translating ribosome affinity purification RNA-seq (TRAP-seq) to determine the translational regulators during nodule organogenesis. Tissue collected from Medicago root 48-hour post-inoculation with rhizobia was used for high throughput sequencing. The correlation between gene expression data from RNA-seq and TRAP-seq indicates the extent of translational regulation during cellular reprogramming in the nodule dedifferentiation process. This sequencing data aided in the identification of several interesting genes. One of these is TAS3, encodes a conserved long noncoding RNA (lncRNA) that produces two alternatively spliced transcripts that are differentially associated with polysomes during bacterial colonization. Another, SKI3, encodes subunit 3 of the SUPERKILLER complex, which participates in mRNA decay, and produces three differentially sized transcripts one of which is translationally upregulated. The functional characterization of SKI3 underlines the significance of this SKI3 protein complex in nodulation as RNAi-induced knock-downs produced significantly fewer nodules. Thus, the TRAP-seq data enhanced the understanding of translational regulation by lncRNA and mRNA decay in Medicago nodule development. (Summary by Suresh Damodaran) Plant Cell 10.1105/tpc.19.00647
The symbiotic relationship between Rhizobium bacteria and leguminous plants like Medicago results in the development of secondary root organs called nodules. The bacteria housed in the nodule infection zone assimilate atmospheric nitrogen for plant growth. In this paper, Traubenik et al. used (RNA-seq) and translating ribosome affinity purification RNA-seq (TRAP-seq) to determine the translational regulators during nodule organogenesis. Tissue collected from Medicago root 48-hour post-inoculation with rhizobia was used for high throughput sequencing. The correlation between gene expression data from RNA-seq and TRAP-seq indicates the extent of translational regulation during cellular reprogramming in the nodule dedifferentiation process. This sequencing data aided in the identification of several interesting genes. One of these is TAS3, encodes a conserved long noncoding RNA (lncRNA) that produces two alternatively spliced transcripts that are differentially associated with polysomes during bacterial colonization. Another, SKI3, encodes subunit 3 of the SUPERKILLER complex, which participates in mRNA decay, and produces three differentially sized transcripts one of which is translationally upregulated. The functional characterization of SKI3 underlines the significance of this SKI3 protein complex in nodulation as RNAi-induced knock-downs produced significantly fewer nodules. Thus, the TRAP-seq data enhanced the understanding of translational regulation by lncRNA and mRNA decay in Medicago nodule development. (Summary by Suresh Damodaran) Plant Cell 10.1105/tpc.19.00647
[altmetric doi=”10.1105/tpc.19.00647″ details=”right” float=”right”]