Repairing a detrimental domestication variant improves tomato harvests
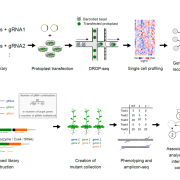
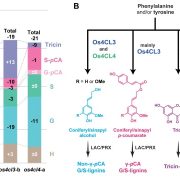
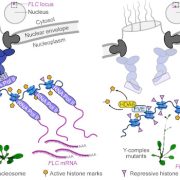
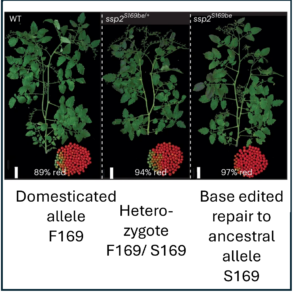 Domesticated plants and animals are remarkable human achievements but were achieved with rather blunt instruments. With the benefit of hindsight, we can now see that some of the genes and alleles that passed through the population bottlenecks and artificial selection process are deleterious. Glaus et al. looked at several tomato wild species, land races, and domesticated varieties. They found large numbers of non-synonymous mutations (coding for a different amino acid) and focused on the genes involved in flowering time, the florigen-activation complex. In tomato, the SELF-PRUNING (SP) gene acts as an anti-florigen that suppresses flowering. Another gene, SUPPRESSOR OF SP (SSP), has previously been used to control plant architecture and time of flowering. The authors identified a related gene, SSP2, that shows a deleterious mutation (S to F) in the DNA-binding domain in domesticated tomatoes. Introgressing the ancestral allele from wild tomatoes led to earlier flowering and a more compact inflorescence. The authors subsequently showed that the deleterious mutation interferes with the ability of SSP2 to bind DNA and act as a transcription factor. Next, they used CRISPR/Cas9 base editing to repair this mutation and restore the ancestral allele in domesticated tomato. The resulting plants showed a more compact growth, earlier flowering, and higher proportion of ripe fruit at harvest. This study shows that today’s powerful genomic tools can identify and repair deleterious alleles that are prevalent in domesticated plants and animals, correcting some unintended outcomes of our ancestors. (Summary by Mary Williams @PlantTeaching.bksy.social @PlantTeaching) Nature Genetics https://doi.org/10.1038/s41588-024-02026-9
Domesticated plants and animals are remarkable human achievements but were achieved with rather blunt instruments. With the benefit of hindsight, we can now see that some of the genes and alleles that passed through the population bottlenecks and artificial selection process are deleterious. Glaus et al. looked at several tomato wild species, land races, and domesticated varieties. They found large numbers of non-synonymous mutations (coding for a different amino acid) and focused on the genes involved in flowering time, the florigen-activation complex. In tomato, the SELF-PRUNING (SP) gene acts as an anti-florigen that suppresses flowering. Another gene, SUPPRESSOR OF SP (SSP), has previously been used to control plant architecture and time of flowering. The authors identified a related gene, SSP2, that shows a deleterious mutation (S to F) in the DNA-binding domain in domesticated tomatoes. Introgressing the ancestral allele from wild tomatoes led to earlier flowering and a more compact inflorescence. The authors subsequently showed that the deleterious mutation interferes with the ability of SSP2 to bind DNA and act as a transcription factor. Next, they used CRISPR/Cas9 base editing to repair this mutation and restore the ancestral allele in domesticated tomato. The resulting plants showed a more compact growth, earlier flowering, and higher proportion of ripe fruit at harvest. This study shows that today’s powerful genomic tools can identify and repair deleterious alleles that are prevalent in domesticated plants and animals, correcting some unintended outcomes of our ancestors. (Summary by Mary Williams @PlantTeaching.bksy.social @PlantTeaching) Nature Genetics https://doi.org/10.1038/s41588-024-02026-9