Remodeling Chromatin in an ARID Environment
The control of gene expression is of fundamental importance for cellular life. In Eukaryotes, linear DNA is wrapped around nucleosomes that constitute a physical barrier to active transcription. Chromatin remodeling complexes modulate the composition, stability and positioning of nucleosomes therefore allowing regulatory factors to access DNA and ultimately regulate gene expression.
Chromatin remodelers are defined by a conserved ATPase domain that fine-tunes DNA-nucleosome interactions (eeviewed in Clapier and Cairns, 2009). The imitation switch (ISWI) family remodelers are characterized by additional SANT and SLIDE domains that bind DNA and histone tails (Boyer et al., 2004). ISWI remodelers form multiple distinct protein complexes and, were shown to move or restructure nucleosomes.
 The plant model organism Arabidopsis thaliana contains two redundant ISWI remodelers, chromatin remodeling 11 and 17 (CHR11/17), which are required for the formation of evenly spaced nucleosomes in gene bodies (Li et al., 2014). CHR11/17 interact with the DTT (DNA binding homeobox and different transcription factors)-domain proteins ringlet 1 and 2 (RLT1/2) and play a major role during plant development (Li et al., 2012). In this issue of The Plant Cell, Tan et al., (2020) identify new CHR11/17 interaction partners and show that these ISWI remodelers are recruited to specific chromatin locations by the DNA/histone binding protein ARID5.
The plant model organism Arabidopsis thaliana contains two redundant ISWI remodelers, chromatin remodeling 11 and 17 (CHR11/17), which are required for the formation of evenly spaced nucleosomes in gene bodies (Li et al., 2014). CHR11/17 interact with the DTT (DNA binding homeobox and different transcription factors)-domain proteins ringlet 1 and 2 (RLT1/2) and play a major role during plant development (Li et al., 2012). In this issue of The Plant Cell, Tan et al., (2020) identify new CHR11/17 interaction partners and show that these ISWI remodelers are recruited to specific chromatin locations by the DNA/histone binding protein ARID5.
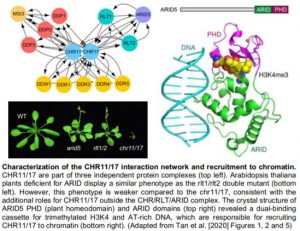
Immunoprecipitation followed by mass spectrometry uncovered a CHR11/17 protein interaction network consisting of three independent complexes (see Figure). Yeast-two hybrid assays revealed direct interactions between RLT1/2 and CHR11/17, between RLT1/2 and ARID5 but not between CHR11/17 and ARID5, suggesting that RLT1/2 act as a bridge between CHR11/17 and ARID5. Consistently, the phenotype of ARID5 loss-of-function plants was similar to the rlt1/rlt2 double mutant (see Figure). In line with additional roles for CHR11/17, outside of the CHR/RLT/ARID complex, the phenotypes of the arid5 and rlt1/2 mutants were not as pronounced as the chr11/17 double mutant.
The ARID5 protein contains an ARID and a PHD (plant homeodomain) domain (see Figure). The crystal structure of these domains revealed a dual-binding cassette binding AT-rich DNA and trimethylated lysine 4 of the histone H3 tail (H3K4me3), respectively. Mutation of the ARID domain or deletion of the PHD domain impaired binding to DNA or H3K4me3 peptides in vitro and, compromised the complementation of the arid5 mutant in vivo. H3K4me3 is a histone post-translational modification marking the transcriptional start site of active genes. Chromatin immunoprecipitation followed by sequencing revealed that ARID5 was enriched at H3K4me3-marked chromatin. This experiment nicely recapitulated in vivo the identified function of the PHD domain as an H3K4me3 reader. Finally, the importance of ARID5 for the recruitment of CHR11/17 to chromatin was illustrated by the reduced binding of CHR11 to two target genes in the absence of ARID5.
In summary, Tan and colleague identified multiple new interaction partners of CHR11/17 and dissected in further detail the roles of RLT1/2 and ARID5 in the recruitment of these remodelers to AT-rich and H3K4me3-marked genes. In the future, it will be interesting to investigate the roles of the two other CHR11/17 complexes and characterize the exact function of CHR11/17 on nucleosome dynamics after their recruitment to specific locations on chromatin.
Sylvain Bischof
Department of Plant and Microbial Biology
University of Zürich, Switzerland
ORCHID ID: 0000-0003-2910-5132
REFERENCES
Lunn, J.E. et al. (2014). Trehalose metabolism in plants. The Plant Journal 79, 544-567.
Lapier, C.R. and Cairns, B.R. (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78, 273-304.
Boyer, L.A. et al. (2004) The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol. 5, 158-63.
Li, G. et al. (2014) ISWI proteins participate in the genome-wide nucleosome distribution in Arabidopsis. Plant J 78, 706-714.
Li, G. et al. (2012) Imitation Switch chromatin remodeling factors and their interacting RINGLET proteins act together in controlling the plant vegetative phase in Arabidopsis. Plant J 72, 261-270.
Tan, L. et al. (2020) Dual recognition of H3K4me3 and DNA by the ISWI component ARID5 regulates floral transition in Arabidopsis. Plant Cell. https://doi.org/10.1105/tpc.19.00944