Pros and cons of various popular RNA sequencing methods
Liu and Zhu et al. systematically compare various methods for investigating transcription.
The Plant Cell: https://doi.org/10.1093/plcell/koad237
By M. Liu and Z. Dong
Guangdong Provincial Key Laboratory of Plant Adaptation and Molecular Design, Guangzhou Key Laboratory of Crop Gene Editing, Innovative Center of Molecular Genetics and Evolution, School of Life Sciences, Guangzhou University, Guangzhou
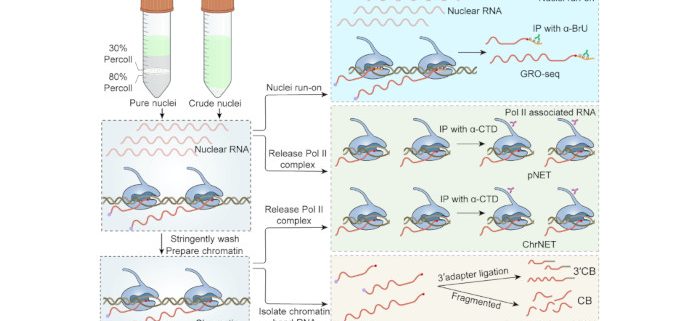
Background: mRNA serves as an intermediate molecule that transfers genetic information from DNA to functional proteins. The level of mRNA expression in a eukaryotic cell, which is determined by processes such as synthesis, processing, and turnover, greatly influences its fate and function. To investigate how a genome produces mRNA through transcription, various methods have been established to directly examine nascent RNA products: 1) native elongation transcripts (NET); 2) global nuclear run-on (GRO); and 3) chromatin-bound RNA as a proxy for nascent RNA (CB). These methods have been used by different labs under various conditions with different pre-treatments and downstream pipelines. Consequently, it remains unclear what the strengths and weaknesses are for each method. This motivated us to conduct a fair comparison using the same plant tissue, pre-treatment, and downstream pipeline whenever possible.
Question: How accurately do these methods reflect transcription? Which method is most suitable for a specific scenario, considering the experimental objectives, feasibility, and budgetary constraints?
Findings: Our findings revealed that the NET and GRO methods performed best when detecting active transcription. CB RNA-seq emerged as a simple yet cost-effective alternative for studying nascent RNA. As an updated version of NET, ChrNET demonstrated higher specificity in capturing nascent RNA, while also reducing sequencing costs. Additionally, 3′CB proved sensitive to transcription-coupled splicing.
Next steps: Moving forward, we plan to leverage the advantages offered by different methods to explore how plants respond at the transcriptional level under external (environmental) and internal (developmental) changes.
Min Liu, Jiafu Zhu, Huijuan Huang, Yan Chen, Zhicheng Dong. (2023). Comparative analysis of nascent RNA sequencing methods and their applications in studies of co-transcriptional splicing dynamics. https://doi.org/10.1093/plcell/koad237
近日,广州大学的刘敏等研究者在The Plant Cell发表了题为Comparative analysis of nascent RNA sequencing methods and their application to co-transcriptional splicing dynamics的研究论文。对转录直接产物——新生RNA进行高通量测序是研究转录的重要手段。在论文中,作者全面系统地比较了几种新生RNA检测方法的优劣特点。
背景回顾:mRNA作为从DNA向功能蛋白传递遗传信息的中间分子,其种类和丰度在很大程度上决定了细胞的命运和功能。在真核细胞中,mRNA的表达量由合成、加工和降解等多个过程共同决定。转录,也就是mRNA生物合成的步骤,因为其重要性受到广泛关注。其中新生RNA测序的方法,不受转录后加工和降解等过程的干扰,比mRNA测序具有更直接的优势,成为研究转录调控的金标准。根据获取新生RNA的生化原理,检测方法分为4类:(1) 通过细胞核run-on在体外亲和标记新生RNA (如GRO-seq);(2) 分离RNA聚合酶II结合的RNA (如NET-seq);(3) 以染色质结合RNA 作为新生RNA的近似代表 (CB RNA-seq);(4) 在体内掺入亲和标记的NTP,富集新生RNA (如TT-seq)。这些方法已被不同的实验室使用,但由于各个实验室种植条件、细胞核提取、下游文库构建等的差异,从新生RNA获取的原理角度,难以比较几种方法的特点。在这项研究中,使用了相同的植物组织、细胞核提取和尽可能相似的实验流程,以确保公平地反映每种方法的长短。
科学问题:寸有所长,尺有所短。哪种(些)方法最逼真地体现细胞内的转录反应?对于特定的研究场景,从实验目标、可行性和成本综合考虑,哪种方法最适合?
研究发现:研究结果显示,NET和GRO类方法在检测基因的活跃转录时效果最好。CB RNA-seq是一个简单且经济实惠的替代方案。作为NET的升级版,ChrNET在捕获新生RNA时具有更高的特异性,同时也降低了测序成本。此外,3′CB能更敏感地检测共转录偶联的剪接事件。
展望未来:下一步打算利用好几种方法的优势,探索植物在转录水平上如何响应对内、外部的环境变化,传递遗传信息。