Plants (RNA) Editors: Testing for Conservation in RNA Editing in moss and angiosperms
When I was a kid, I remember keeping a bottle of wite-out with my pens for the inevitable spelling mistake. Now, in the digital age, I either let my word processor autocorrect spelling errors or wait for the red squiggly line under suspicious words.
On any given day, a single (plant) cell by far out-writes me. They truly are prolific writers, copying DNA and RNA endlessly. But sometimes, a mutation sneaks into the genome that will need correction in the RNA copy. Just like us, they suffer the consequences of bad spelling. However, cells do not use white paint to cover up the error: they use RNA editors called pentatricopeptide repeat (PPR) proteins. Here, Oldenkott et al (2020) offer us a glimpse into the selective pressures faced by plants to correct their organellar spelling mistakes.
 PPR proteins bear between 2 and 30 repeats, each exhibiting specificity toward a given ribonucleotide that together dictate the RNA sequence motif the PPR protein will recognize. The most critical positions within each repeat are the 5th and last amino acids, as they largely inform to which ribonucleotide their PPR will bind. This fact was exploited to decipher the PPR code, effectively predicting the RNA target of a given PPR protein (Barkan et al., 2012). RNA editors do not bind to the error site itself, but rather upstream of it. A DYW domain that is either part of the same protein or recruited then catalyzes the conversion of the wrong base to correct the error.
PPR proteins bear between 2 and 30 repeats, each exhibiting specificity toward a given ribonucleotide that together dictate the RNA sequence motif the PPR protein will recognize. The most critical positions within each repeat are the 5th and last amino acids, as they largely inform to which ribonucleotide their PPR will bind. This fact was exploited to decipher the PPR code, effectively predicting the RNA target of a given PPR protein (Barkan et al., 2012). RNA editors do not bind to the error site itself, but rather upstream of it. A DYW domain that is either part of the same protein or recruited then catalyzes the conversion of the wrong base to correct the error.
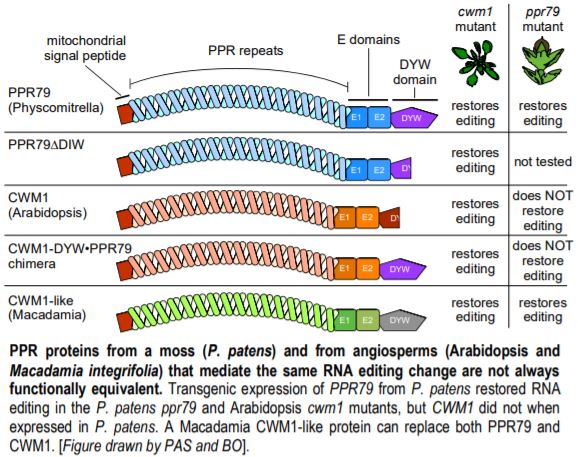
The authors were interested in the evolution of PPR proteins, but a full picture is only possible when also looking at the associated site they edit. The authors therefore combed through the vast sequence space in the moss Physcomitrium (Physcomitrella) patens, and the angiosperms Arabidopsis (Arabidopsis thaliana) and Macadamia (Macadamia integrifolia; yes, the tasty nuts). They discovered that mitochondrial nad5 transcripts shared one RNA editing site in all three species. Two of the PPR proteins targeting this site are known: PPR79 in P. patens, with a full DYW domain, and CWM1 (CELL WALL MAINTAINER 1) in Arabidopsis, with a truncated (and non-functional) DYW domain (see Figure) (Uchida et al., 2011; Hu et al., 2016). The RNA sequence upstream of the edited site was also highly conserved, prompting the authors to test whether PPR79 could substitute for CWM1 (and vice versa).
Although PPR79 and CWM1 only share 20-25% identity, the authors discovered that PPR79 was able to properly edit the nad5 site when expressed in the Arabidopsis cwm1 mutant. However, the converse was not true: CWM1 expressed in the P. patens ppr79 mutant failed to restore editing, even when the full DYW domain from PPR79 was added to CWM1. The CWM1-like protein from Macadamia, another flowering plant, was also able to properly edit the nad5 site when expressed in the ppr79 and cwm1 mutants (see Figure).
Efficient editing by CWM1 in Arabidopsis needs the recruitment of a protein with a DYW domain to catalyze ribonucleotide exchange in trans. Because the P. patens editing system is much simpler than the Arabidopsis one, the authors hypothesize that CWM1 cannot recruit this partner in P. patens. Surprisingly, even though PPR79 comes with its own DYW domain, a truncated form of PPR79 can recruit a DYW domain when expressed in Arabidopsis.
With these results, the authors provide a system to systematically test the evolution of PPR proteins and RNA editing systems. Their results might even answer this burning question: how did plants survive their first spelling error?
Patrice A. Salomé
Science Editor
ORCID: 0000-0003-4452-9064
References:
Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond C.S, Small I (2012). A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins. PLoS Genet. 8 e1002910.
Hu Z., Vanderhaeghen R, Cools T, Wang Y, De Clercq I, Leroux O, Nguyen L, Belt K, Millar AH, Audenaert D, Hilson P, Small I, Mouille G, Vernhettes S, Van BF, Whelan J, Hofte H, and De Veylder L (2016). Mitochondrial Defects Confer Tolerance against Cellulose Deficiency. Plant Cell 28:2276-2290.
Oldenkott B, Burger M, Hein A-C, Jörg A, Senkler J, Braun H-P, Knoop V, Takenaka M, Schallenberg-Rüdinger M (2020). One C-to-U RNA editing site and two independently evolved editing factors: testing reciprocal complementation with DYW-type PPR proteins from the moss Physcomitrium (Physcomitrella) patens and the flowering plants Macadamia integrifolia and Arabidopsis thaliana. Published June 2020. DOI: https://doi.org/10.1105/tpc.20.00311
Uchida M, Ohtani S, Ichinose M, Sugita C, and Sugita M (2011). The PPR-DYW proteins are required for RNA editing of rps14, cox1 and nad5 transcripts in Physcomitrella patens mitochondria. FEBS Lett. 585:2367-2371.