Plant Science Research Weekly: September 9, 2022
Removing systemic barriers to equity, diversity, and inclusion: Report of the 2019 Plant Science Research Network workshop “Inclusivity in the Plant Sciences”
 In January 2019, a workshop was convened to explore strategies to address inclusivity in the plant sciences, and the recommendations emerging from this workshop are presented in a new report by Henkhaus et al. The authors recognize that this is an expansive challenge and requires action on many fronts, which they group into four areas: Early STEM exposure, Representation through personal stories, Shifting mentoring and sponsorship norms to broaden participation, and Breaking down systemic and institutional barriers. Specific recommendations and challenges for each area are provided. For example, ensuring that people are supported as they transition between degree programs or jobs is important, but how can that be implemented? Outreach by researchers into the broader community is crucial, but rarely rewarded by tenure or salary decisions. Science benefits when researchers are free to work in different countries, but funding and immigration policies are tremendous barriers to effective international collaboration and cross-pollination. And finally, when people in power have benefitted from current systems, how can they be motivated to use that power to develop new, more inclusive systems? Included in the Supplemental Information are eight potential pilot projects envisioned by the workshop participants. (Summary by Mary Williams @PlantTeaching) Plant Direct 10.1002/pld3.432
In January 2019, a workshop was convened to explore strategies to address inclusivity in the plant sciences, and the recommendations emerging from this workshop are presented in a new report by Henkhaus et al. The authors recognize that this is an expansive challenge and requires action on many fronts, which they group into four areas: Early STEM exposure, Representation through personal stories, Shifting mentoring and sponsorship norms to broaden participation, and Breaking down systemic and institutional barriers. Specific recommendations and challenges for each area are provided. For example, ensuring that people are supported as they transition between degree programs or jobs is important, but how can that be implemented? Outreach by researchers into the broader community is crucial, but rarely rewarded by tenure or salary decisions. Science benefits when researchers are free to work in different countries, but funding and immigration policies are tremendous barriers to effective international collaboration and cross-pollination. And finally, when people in power have benefitted from current systems, how can they be motivated to use that power to develop new, more inclusive systems? Included in the Supplemental Information are eight potential pilot projects envisioned by the workshop participants. (Summary by Mary Williams @PlantTeaching) Plant Direct 10.1002/pld3.432
ABP1 (it’s back!) binds auxin and signals fast auxin responses
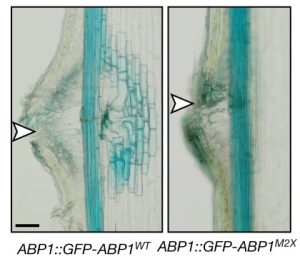 One of the longstanding questions in plant biology concernes how auxin coordinates both gene expression in the nucleus and the so-called fast responses at the cell periphery including proton extrusion and cytoskeletal rearrangements. At this point the nuclear branch is well understood, but the cell periphery responses have proven to be be problematic. Fifty years ago, membrane fractions with strong and specific binding to auxin were identified, with the Auxin Binding Protein (ABP) subsequently purified. Since then, ABP’s role has been contentious, most notably with the finding that the phenoytype of the Arabidopsis abp1 mutant is a consequence of a nearby but unrelated mutation. Happily, ABP1 has had another turn of fortune with the publication by Friml et al. of definitive data showing that ABP1 is indeed an auxin-binding protein that signals some of the fast-auxin responses. The authors show that ABP1 binds to auxin and resides both in the ER (known previously, as it has an ER-targeting KDEL sequence) and within the apoplast. At the cell surface, ABP1 interacts with a membrane-localized protein kinase, TMK1. In the presence of auxin, this complex phosphorylates thousands of proteins including plasma-membrane H+-ATPases and myosin/myosin-binding proteins. Using a series of ABP1 mutants including ones that fail to bind auxin, the authors show that auxin+ABP1 and TMK1 are needed for auxin canalization responses in shoot de novo and wound-induced vasculature formation. However, other non-transcriptional responses including calcium transients and root growth inhibition do not appear to be mediated by ABP1; additionally, notable phenotypic differences between abp1 and tmk1 mutants demonstrate that that more chapters in the ABP1 story await discovery. (Summary by Mary Williams @PlantTeaching) Nature 10.1038/s41586-022-05187-x
One of the longstanding questions in plant biology concernes how auxin coordinates both gene expression in the nucleus and the so-called fast responses at the cell periphery including proton extrusion and cytoskeletal rearrangements. At this point the nuclear branch is well understood, but the cell periphery responses have proven to be be problematic. Fifty years ago, membrane fractions with strong and specific binding to auxin were identified, with the Auxin Binding Protein (ABP) subsequently purified. Since then, ABP’s role has been contentious, most notably with the finding that the phenoytype of the Arabidopsis abp1 mutant is a consequence of a nearby but unrelated mutation. Happily, ABP1 has had another turn of fortune with the publication by Friml et al. of definitive data showing that ABP1 is indeed an auxin-binding protein that signals some of the fast-auxin responses. The authors show that ABP1 binds to auxin and resides both in the ER (known previously, as it has an ER-targeting KDEL sequence) and within the apoplast. At the cell surface, ABP1 interacts with a membrane-localized protein kinase, TMK1. In the presence of auxin, this complex phosphorylates thousands of proteins including plasma-membrane H+-ATPases and myosin/myosin-binding proteins. Using a series of ABP1 mutants including ones that fail to bind auxin, the authors show that auxin+ABP1 and TMK1 are needed for auxin canalization responses in shoot de novo and wound-induced vasculature formation. However, other non-transcriptional responses including calcium transients and root growth inhibition do not appear to be mediated by ABP1; additionally, notable phenotypic differences between abp1 and tmk1 mutants demonstrate that that more chapters in the ABP1 story await discovery. (Summary by Mary Williams @PlantTeaching) Nature 10.1038/s41586-022-05187-x
Two fern genome papers
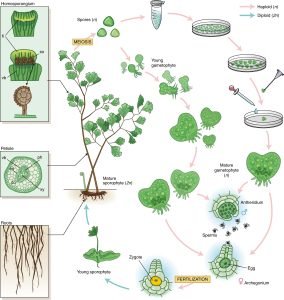 The September 2022 issue of Nature Plants includes two papers describing the analysis of homosporous fern genomes. Ferns are interesting for many reasons including the diversity of the clade, their typically very large genomes, their often free-living gametophytes, and in many cases, homospory (a single type of spore), which is distinct from angiosperm’s heterospory (separate male and female spores). The first of these papers, by Marchant, Chen, and Cai et al., describes the “Dynamic genome evolution in a model fern.” Ceratopteris richardii, also known as C-fern, has been a useful model system in many classrooms (https://c-fern.org/). The authors explored the presence of genes associated with flowering and reproduction in this non-flowering plant, as well as genes derived from horizontal gene transfer and genes that might contribute to medicinal properties. The second paper explores the genome of maiden hair fern, Adiantum capillus-veneris L. Key findings from this work include the phylogenetic analysis of genes that are involved in seed desiccation tolerance and genes involved in defense responses and leaf development. (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-022-01226-7 and 10.1038/s41477-022-01222-x.
The September 2022 issue of Nature Plants includes two papers describing the analysis of homosporous fern genomes. Ferns are interesting for many reasons including the diversity of the clade, their typically very large genomes, their often free-living gametophytes, and in many cases, homospory (a single type of spore), which is distinct from angiosperm’s heterospory (separate male and female spores). The first of these papers, by Marchant, Chen, and Cai et al., describes the “Dynamic genome evolution in a model fern.” Ceratopteris richardii, also known as C-fern, has been a useful model system in many classrooms (https://c-fern.org/). The authors explored the presence of genes associated with flowering and reproduction in this non-flowering plant, as well as genes derived from horizontal gene transfer and genes that might contribute to medicinal properties. The second paper explores the genome of maiden hair fern, Adiantum capillus-veneris L. Key findings from this work include the phylogenetic analysis of genes that are involved in seed desiccation tolerance and genes involved in defense responses and leaf development. (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-022-01226-7 and 10.1038/s41477-022-01222-x.
Teff breeding potentials from data-driven, participatory characterization of farmer varieties
 There is a clear need to synergize advances from cutting-edge genomic approaches with the needs and knowledge of growers, particularly small-holder growers who have access to much of a crop’s genetic diversity. Here, Woldeyohannes, Iohannes et al. took a transdisciplinary approach to explore the breeding potential of teff (Eragrostis tef), a cereal crop widely grown in Ethiopia which is also considered a neglected and underutilized species (NUS). (The spongy flat bread served with Ethiopian food is made from teff). The authors assembled a collection of 366 teff varieties (321 farmer varieties derived from landraces and 45 improved lines), which they characterized by genome-wide molecular markers and by evaluation by a panel of experienced teff growers (15 women and 20 men), providing a meaningful connection between genetic analysis and farmer preferences. The study found that variance for production traits and farmer preferences had both geographic and genetic correlations. GWAS analysis identified loci and candidate genes related to phenology, yield, local adaptation, and farmers’ appreciation. Interestingly, men and women evaluated the crops somewhat differently, which has been shown previously in smallholder farmers in Africa, and “may reflect gender roles in the value chain” (women are often more interested in use traits, and men in agronomic traits such as plant height). This study provides an exciting model in which to identify avenues to improve neglected and underutilized species in ways that will be beneficial to those who grow them. (Summary by Mary Williams @PlantTeaching) eLIFE 10.7554/eLife.80009
There is a clear need to synergize advances from cutting-edge genomic approaches with the needs and knowledge of growers, particularly small-holder growers who have access to much of a crop’s genetic diversity. Here, Woldeyohannes, Iohannes et al. took a transdisciplinary approach to explore the breeding potential of teff (Eragrostis tef), a cereal crop widely grown in Ethiopia which is also considered a neglected and underutilized species (NUS). (The spongy flat bread served with Ethiopian food is made from teff). The authors assembled a collection of 366 teff varieties (321 farmer varieties derived from landraces and 45 improved lines), which they characterized by genome-wide molecular markers and by evaluation by a panel of experienced teff growers (15 women and 20 men), providing a meaningful connection between genetic analysis and farmer preferences. The study found that variance for production traits and farmer preferences had both geographic and genetic correlations. GWAS analysis identified loci and candidate genes related to phenology, yield, local adaptation, and farmers’ appreciation. Interestingly, men and women evaluated the crops somewhat differently, which has been shown previously in smallholder farmers in Africa, and “may reflect gender roles in the value chain” (women are often more interested in use traits, and men in agronomic traits such as plant height). This study provides an exciting model in which to identify avenues to improve neglected and underutilized species in ways that will be beneficial to those who grow them. (Summary by Mary Williams @PlantTeaching) eLIFE 10.7554/eLife.80009
The vacuolar H+/Ca transporter CAX1 participates in submergence and anoxia stress responses
 Calcium (Ca) signaling is one of the primary plant responses to confront abiotic stresses. To coordinate Ca levels in the different compartments of the cell, several transporters are needed including the H+/Ca exchangers (CAXs) in the tonoplast (vacuolar membrane). While the role of calcium signaling has been studied for osmotic and salt stresses in plants, little is known about the role of calcium in responding to submergence stress. Submergence leads to several stresses, from limited light availability to the oxidative stress produced after hypoxic periods. In this work, Yang et al. analyzed CAX1, an exchanger that pumps Ca from the cytoplasm to the vacuole. The authors described that in Arabidopsis thaliana, cax1 mutants show an exceptional tolerance to submergence and anoxic stresses, resembling plants primed in advance for facing these stresses. Post-anoxia, lower levels of Reactive Oxygen Species (ROS) were found in cax1 lines. Finally, it was also demonstrated that cax1 line showed a different Ca signature during reoxygenation than control plants. Overall, this is one of the first works that shows how a tonoplast localized H+/Ca transporter can influence anoxia tolerance in plants. (Summary by Eva María Gómez Álvarez, @eva_ga96). Plant Physiol. 10.1093/plphys/kiac375
Calcium (Ca) signaling is one of the primary plant responses to confront abiotic stresses. To coordinate Ca levels in the different compartments of the cell, several transporters are needed including the H+/Ca exchangers (CAXs) in the tonoplast (vacuolar membrane). While the role of calcium signaling has been studied for osmotic and salt stresses in plants, little is known about the role of calcium in responding to submergence stress. Submergence leads to several stresses, from limited light availability to the oxidative stress produced after hypoxic periods. In this work, Yang et al. analyzed CAX1, an exchanger that pumps Ca from the cytoplasm to the vacuole. The authors described that in Arabidopsis thaliana, cax1 mutants show an exceptional tolerance to submergence and anoxic stresses, resembling plants primed in advance for facing these stresses. Post-anoxia, lower levels of Reactive Oxygen Species (ROS) were found in cax1 lines. Finally, it was also demonstrated that cax1 line showed a different Ca signature during reoxygenation than control plants. Overall, this is one of the first works that shows how a tonoplast localized H+/Ca transporter can influence anoxia tolerance in plants. (Summary by Eva María Gómez Álvarez, @eva_ga96). Plant Physiol. 10.1093/plphys/kiac375
Modus operandi of Nod-independent symbiosis in Aeschynomene evenia
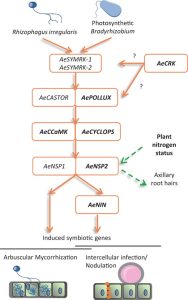 Symbioses and nodule organogenesis processes that occur independently of Nod-factors are relatively unexplored. Quilbe et al. investigated the semi-aquatic legume Aeschynomene evenia, which has recently been shown to establish symbioses with Bradyrhizobium sp. that do not produce Nod factors. The authors used an EMS mutagenesis approach to generate mutations in genes known to be involved in the Nod-factor symbiotic pathway: AePOLLUX, Ca2+/calmodulin dependent kinase (AeCCaMK), AeCYCLOPS, nodulation signaling pathway 2 (AeNSP2), and nodule inception (NIN), as well as a symbiosis-associated cysteine-rich receptor-like kinase (AeCRK). Nodulation kinetics and early nodulin gene expression of the symbiosis mutants were significantly altered in comparison to wild type. For example, pollux-2, pollux-3, pollux-4, cyclops-2 and nin mutants (except nin1) did not produce nodules, while nin-1, pollux-1 and pollux-5 produced underdeveloped nodules with low or no nitrogenase activity. Mycorrhizal colonization and expression of mycorrhization markers were truncated in the roots of the pollux-2, ccamk-3, and cyclops-2 mutants. Axillary root hair crowns, the site of bradyrhizobia colonization, were completely compromised in nsp2 mutants. High nitrate supply suppressed axillary root development and symbiosis associated transcription, while this “nitrogen effect” was lost in nsp2 mutants. These findings show that in Aeschynomene evenia, there is a similar involvement of the Nod signaling pathway throughout the process of rhizobial invasion and symbiosome formation as in other Nod-dependent legumes. (Summary by Lekshmy Sathee @lekshmysnair) Plant Physiol. 10.1093/plphys/kiac325
Symbioses and nodule organogenesis processes that occur independently of Nod-factors are relatively unexplored. Quilbe et al. investigated the semi-aquatic legume Aeschynomene evenia, which has recently been shown to establish symbioses with Bradyrhizobium sp. that do not produce Nod factors. The authors used an EMS mutagenesis approach to generate mutations in genes known to be involved in the Nod-factor symbiotic pathway: AePOLLUX, Ca2+/calmodulin dependent kinase (AeCCaMK), AeCYCLOPS, nodulation signaling pathway 2 (AeNSP2), and nodule inception (NIN), as well as a symbiosis-associated cysteine-rich receptor-like kinase (AeCRK). Nodulation kinetics and early nodulin gene expression of the symbiosis mutants were significantly altered in comparison to wild type. For example, pollux-2, pollux-3, pollux-4, cyclops-2 and nin mutants (except nin1) did not produce nodules, while nin-1, pollux-1 and pollux-5 produced underdeveloped nodules with low or no nitrogenase activity. Mycorrhizal colonization and expression of mycorrhization markers were truncated in the roots of the pollux-2, ccamk-3, and cyclops-2 mutants. Axillary root hair crowns, the site of bradyrhizobia colonization, were completely compromised in nsp2 mutants. High nitrate supply suppressed axillary root development and symbiosis associated transcription, while this “nitrogen effect” was lost in nsp2 mutants. These findings show that in Aeschynomene evenia, there is a similar involvement of the Nod signaling pathway throughout the process of rhizobial invasion and symbiosome formation as in other Nod-dependent legumes. (Summary by Lekshmy Sathee @lekshmysnair) Plant Physiol. 10.1093/plphys/kiac325
Identification of a cell signaling cascade that regulate s broad-spectrum resistance
s broad-spectrum resistance
Many different pathogens attack plants, and many genes have been identified that confer pathogen-specific resistance. In a recent study, Hao et al. identified a signaling cascade that regulates broad-spectrum disease resistance. This pathway is composed of a rice ubiquitin ligase OsPUB73, a VQ motif-containing protein OsVQ25, and a transcription factor OsWRKY53. As is characteristic of VQ-motif proteins, OsVQ25 interacts with OsWRKY53 and through this interaction suppresses OsWRKY53-mediated induction of defense genes. Upstream, OsPUB73 interacts with OsVQ25 and causes its proteasomal degradation. The authors showed that OsPUB73 is positive regulator of broad-spectrum immunity, as knockout plants are more susceptible to both the blight bacterium Xanthomonas oryzae pv. oryzae (Xoo) and the rice blast fungus Magnaporthe oryzae. By contrast, OsVQ25 is a negative regulator of immunity, as enhanced resistance against both pathogens was observed in osvq25 mutants. Additionally, this enhanced broad-spectrum resistance comes without any observable effects on major agronomic traits such as plant height, kernels per spike, or grain weight. Apparently, induction of immunity involves OsPUB73-mediated degradation of OsVQ25 leading to the release of OsWKRY53 to enhance expression of defense-related genes. Summary by Kamal Kumar Malukani, @KamalMalukani. Cell Reports, 10.1016/j.celrep.2022.111235
Agarwood harnesses hornets for rapid dispersal of non-dormant seeds
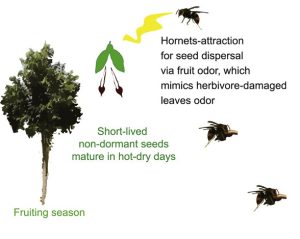 The seeds of the Agarwood tree (Aquilaria sinensis) lose viability within a few hours of the fruit splitting, so need to reach the ground as soon as possible. Qin et al. report that agarwood achieves this by fooling hornets (Vespa spp.), which generally eat herbivore insects such as caterpillars. When plants are attacked by caterpillars, they release various volatile organic compounds (VOCs) as danger signals. Hornets associate these VOCs with the presence of herbivore insects and get attracted to the source of the VOCs where they find and eat the caterpillars. The authors report that when agarwood fruit splits, it starts to release a similar set of VOCs that are released during caterpillar attack. Additionally, the hanging seed mimics a hanging caterpillar. The hornets, attracted by the VOCs, remove the seed from the plant and take them to its nest, generally on other trees. Hornets consume only the elaiosome (a nutrient-rich structure often found on insect-dispersed seeds) and drop the rest of the seed, often in the shade of other trees which facilitate survival and germination of these desiccation-sensitive non-dormant seeds. (Summary by Kamal Kumar Malukani @KamalMalukani) Curr. Biol. 10.1016/j.cub.2022.06.034
The seeds of the Agarwood tree (Aquilaria sinensis) lose viability within a few hours of the fruit splitting, so need to reach the ground as soon as possible. Qin et al. report that agarwood achieves this by fooling hornets (Vespa spp.), which generally eat herbivore insects such as caterpillars. When plants are attacked by caterpillars, they release various volatile organic compounds (VOCs) as danger signals. Hornets associate these VOCs with the presence of herbivore insects and get attracted to the source of the VOCs where they find and eat the caterpillars. The authors report that when agarwood fruit splits, it starts to release a similar set of VOCs that are released during caterpillar attack. Additionally, the hanging seed mimics a hanging caterpillar. The hornets, attracted by the VOCs, remove the seed from the plant and take them to its nest, generally on other trees. Hornets consume only the elaiosome (a nutrient-rich structure often found on insect-dispersed seeds) and drop the rest of the seed, often in the shade of other trees which facilitate survival and germination of these desiccation-sensitive non-dormant seeds. (Summary by Kamal Kumar Malukani @KamalMalukani) Curr. Biol. 10.1016/j.cub.2022.06.034



