Plant Science Research Weekly: September 11, 2020
Review. Roles of plant retinoblastoma protein: cell cycle and beyond
 The cell cycle is at the heart of processes such as cell division, fate acquisition and cell cycle exit towards differentiation. Decades of cell cycle research in animals and yeast have outlined the main components that control the cycle’s transitions, such as cyclins, cyclin-dependent kinases, E2F transcription factors and the Retinoblastoma (RB1) tumor suppressor. Mostly studied in mammalian models, RB1 is a pivotal protein that acts through different complexes to regulate cell cycle transitions at G1/S and G2/M phases. In plants, several lines of research during the mid-1990s led to discovery that the function and modules of core cell cycle regulators, including Retinoblastoma-related (RBR1) proteins, were conserved in plants and unicellular algae, however, new evidence implicates RBR1 in a plethora of novel processes that go past its canonical cell cycle function. A recent review by Desvoyes & Gutierrez describes the multiple roles of RBR1, which acts as a molecular hub coordinating growth, proliferation and differentiation via the formation of multimeric complexes tuned by many regulatory layers, e.g., phosphorylation, ubiquitination, and upstream inhibitors. The authors illustrate that RBR1 interacts with cell lineage-specific transcription factors and chromatin remodeling components through different regions and motifs to modulate several processes, such as cell cycle transcriptional control, cell differentiation in the root apical meristem and the stomatal lineage, as well as embryo and gametophyte development. Moreover, the authors discuss evidence that RBR1 seems to have a role in DNA-damage response and pathogen infection, although a mechanistic view for RBR1 role in these processes is still lacking. This fresh review highlights RBR1 participation in multiple processes and spotlights promising research avenues: how does phosphorylation modulate RBR1 interactions and what complexes might RBR1 be forming with transcriptional factors belonging to other families? (Summary by Jesus Leon @jesussaur) EMBO J. 10.15252/embj.2020105802
The cell cycle is at the heart of processes such as cell division, fate acquisition and cell cycle exit towards differentiation. Decades of cell cycle research in animals and yeast have outlined the main components that control the cycle’s transitions, such as cyclins, cyclin-dependent kinases, E2F transcription factors and the Retinoblastoma (RB1) tumor suppressor. Mostly studied in mammalian models, RB1 is a pivotal protein that acts through different complexes to regulate cell cycle transitions at G1/S and G2/M phases. In plants, several lines of research during the mid-1990s led to discovery that the function and modules of core cell cycle regulators, including Retinoblastoma-related (RBR1) proteins, were conserved in plants and unicellular algae, however, new evidence implicates RBR1 in a plethora of novel processes that go past its canonical cell cycle function. A recent review by Desvoyes & Gutierrez describes the multiple roles of RBR1, which acts as a molecular hub coordinating growth, proliferation and differentiation via the formation of multimeric complexes tuned by many regulatory layers, e.g., phosphorylation, ubiquitination, and upstream inhibitors. The authors illustrate that RBR1 interacts with cell lineage-specific transcription factors and chromatin remodeling components through different regions and motifs to modulate several processes, such as cell cycle transcriptional control, cell differentiation in the root apical meristem and the stomatal lineage, as well as embryo and gametophyte development. Moreover, the authors discuss evidence that RBR1 seems to have a role in DNA-damage response and pathogen infection, although a mechanistic view for RBR1 role in these processes is still lacking. This fresh review highlights RBR1 participation in multiple processes and spotlights promising research avenues: how does phosphorylation modulate RBR1 interactions and what complexes might RBR1 be forming with transcriptional factors belonging to other families? (Summary by Jesus Leon @jesussaur) EMBO J. 10.15252/embj.2020105802
Letter. Planting equity: Using what we know to cultivate growth as a plant biology community
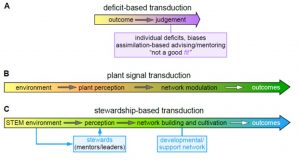 COVID-19 has derailed everyone’s lives and plans; 2020 is truly an “annus horribilis.” Yet clearly this pandemic has hit some harder than others. Note for example the greatly disproportionate mortality in Black and Indigenous populations, which surely contributed to the outpouring of outrage following the murders of George Floyd, Breonna Taylor and others. For many, this year has been a tipping point in terms of recognizing the impact of systemic racism and how it permeates and influences everything, even science. In this Letter, Beronda Montgomery addresses “ways we might use our knowledge as plant scientists to help us address systemic racism and persistent issues of inequity in scientific spaces and far beyond.” She makes a compelling analogy between our understanding of how the environment affects a plant’s phenotype, and how the environment in which a scientist works affects their success. She urges us to move from a “deficit” based model, in which a lack of success is attributed to an individual’s own failings, to a “stewardship” based model, and work towards “building, cultivating, and sustaining an equitable community in plant biology.” What does this entail? It requires a “shift from “gatekeeping” practices in which we carefully guard the gates and make subjective, and often biased, decisions about who “fits” and is worthy to enter, to actively using “groundskeeping” practices, whereby we thoughtfully tend our environments to actively support access and progress, and recognize and remove barriers to individual success.” (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1105/tpc.20.00589
COVID-19 has derailed everyone’s lives and plans; 2020 is truly an “annus horribilis.” Yet clearly this pandemic has hit some harder than others. Note for example the greatly disproportionate mortality in Black and Indigenous populations, which surely contributed to the outpouring of outrage following the murders of George Floyd, Breonna Taylor and others. For many, this year has been a tipping point in terms of recognizing the impact of systemic racism and how it permeates and influences everything, even science. In this Letter, Beronda Montgomery addresses “ways we might use our knowledge as plant scientists to help us address systemic racism and persistent issues of inequity in scientific spaces and far beyond.” She makes a compelling analogy between our understanding of how the environment affects a plant’s phenotype, and how the environment in which a scientist works affects their success. She urges us to move from a “deficit” based model, in which a lack of success is attributed to an individual’s own failings, to a “stewardship” based model, and work towards “building, cultivating, and sustaining an equitable community in plant biology.” What does this entail? It requires a “shift from “gatekeeping” practices in which we carefully guard the gates and make subjective, and often biased, decisions about who “fits” and is worthy to enter, to actively using “groundskeeping” practices, whereby we thoughtfully tend our environments to actively support access and progress, and recognize and remove barriers to individual success.” (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1105/tpc.20.00589
The vitamin B1 family grows: detection and roles of thiamine triphosphate in plants
 The term “vitamin B1” indicates a family of molecules that includes thiamine, thiamine monophosphate and thiamine diphosphate (TDP). The latter is the most famous family member and plays a pivotal role in carbon metabolism. Thanks to an optimized detection protocol, Hofmann and colleagues have demonstrated that Arabidopsis thaliana shoots also produce thiamine triphosphate (TTP). The production of TTP is likely due to the proton motive force in mitochondria and/or plastids and TDP is its most likely precursor. The accumulation of TTP increases with light intensity and peaks during the day, but is independent of the circadian clock. It rather reflects the biosynthesis of TDP, which is a photosynthesis-driven process that takes place in green tissues. Because of the concomitant accumulation of TDP and TTP, the authors hypothesize that conversion of TDP to TTP may serve to dissipate excess free TDP, especially in the plastids and/or mitochondria. In analogy with what has been recently observed in mammals, plant TTP could also modulate important enzymes involved in amino acid biosynthesis. (Summary by Elisa Dell’Aglio) Plant Direct 10.1002/pld3.258
The term “vitamin B1” indicates a family of molecules that includes thiamine, thiamine monophosphate and thiamine diphosphate (TDP). The latter is the most famous family member and plays a pivotal role in carbon metabolism. Thanks to an optimized detection protocol, Hofmann and colleagues have demonstrated that Arabidopsis thaliana shoots also produce thiamine triphosphate (TTP). The production of TTP is likely due to the proton motive force in mitochondria and/or plastids and TDP is its most likely precursor. The accumulation of TTP increases with light intensity and peaks during the day, but is independent of the circadian clock. It rather reflects the biosynthesis of TDP, which is a photosynthesis-driven process that takes place in green tissues. Because of the concomitant accumulation of TDP and TTP, the authors hypothesize that conversion of TDP to TTP may serve to dissipate excess free TDP, especially in the plastids and/or mitochondria. In analogy with what has been recently observed in mammals, plant TTP could also modulate important enzymes involved in amino acid biosynthesis. (Summary by Elisa Dell’Aglio) Plant Direct 10.1002/pld3.258
Real time quantitative imaging of transcriptional activity at the single cell level
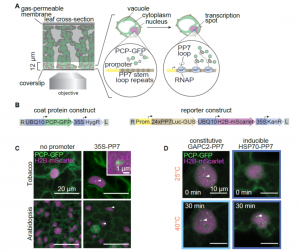 How plants transcriptionally operate developmental programs and responses to stress in time and space has been an important question in plant biology. Fluorescent protein reporters are commonly used to address this question, but their performance is limited at short timescales (<30 min) before the proteins get matured. Also, expression of fluorescent proteins can be influenced by various processes before and after maturation (e.g., RNA transport and protein degradation). To overcome these limitations, Alamos et al. implemented an mRNA fluorescent-tagging approach in Nicotiana benthamiana and Arabidopsis thaliana. In this system, the promoter of a gene is tagged with the PP7 (or MS2) viral DNA sequence that when transcribed accumulates fluorescent proteins already expressed in the nucleus. The signal can be immediately detected as a fluorescent spot, and the signal intensity reports the absolute count of RNA polymerase molecules actively transcribing the target gene. The authors generated stably transformed lines of Arabidopsis with construct targeting the heat shock protein HSP101 to monitor plant responses to heat stress. They found that only a fraction of cells was transcriptionally responsive to the stress, and among responsive cells, instantaneous transcription events were detected in a heterogeneous manner. The tissue-wide transcription rate of the heat-responsive gene was controlled by a switch-like mechanism where heat stress induces the probability that individual cells engage in transcription, whereas the transcription level of already actively transcribing cells remained stable. Finally, they showed that the cellular heterogeneity in the transcriptional response to heat shock is mainly explained by stochastic processes at the level of each allele rather than cells having a different chemical composition. This approach opens the door for single cell interrogation of transcription variability and dynamics in plants. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2020.08.30.274621
How plants transcriptionally operate developmental programs and responses to stress in time and space has been an important question in plant biology. Fluorescent protein reporters are commonly used to address this question, but their performance is limited at short timescales (<30 min) before the proteins get matured. Also, expression of fluorescent proteins can be influenced by various processes before and after maturation (e.g., RNA transport and protein degradation). To overcome these limitations, Alamos et al. implemented an mRNA fluorescent-tagging approach in Nicotiana benthamiana and Arabidopsis thaliana. In this system, the promoter of a gene is tagged with the PP7 (or MS2) viral DNA sequence that when transcribed accumulates fluorescent proteins already expressed in the nucleus. The signal can be immediately detected as a fluorescent spot, and the signal intensity reports the absolute count of RNA polymerase molecules actively transcribing the target gene. The authors generated stably transformed lines of Arabidopsis with construct targeting the heat shock protein HSP101 to monitor plant responses to heat stress. They found that only a fraction of cells was transcriptionally responsive to the stress, and among responsive cells, instantaneous transcription events were detected in a heterogeneous manner. The tissue-wide transcription rate of the heat-responsive gene was controlled by a switch-like mechanism where heat stress induces the probability that individual cells engage in transcription, whereas the transcription level of already actively transcribing cells remained stable. Finally, they showed that the cellular heterogeneity in the transcriptional response to heat shock is mainly explained by stochastic processes at the level of each allele rather than cells having a different chemical composition. This approach opens the door for single cell interrogation of transcription variability and dynamics in plants. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2020.08.30.274621
Single nucleus analysis of Arabidopsis seeds reveals new cell types and imprinting dynamics
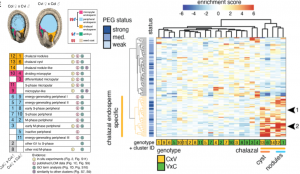 Arabidopsis seeds consist of various tissue like seed coat, embryo, and endosperm. The endosperm provides the nutrient supplies to the growing embryo and has three domains namely, micropylar (surrounding embryo), chalazal (opposite end of the seed), and peripheral (in between micropylar and chalazal) domain. Even though there is transcriptomic data available for these different domains of endosperm, it is not yet known whether there is cell-type heterogeneity within these domains. In this paper Picard et al. carried out single nucleus RNA-sequencing (snRNA-Seq) on F1 seeds from a reciprocal cross between the Columbia (Col-0) and Cape Verde Islands (Cvi-0) to identify cell-type and imprinting heterogeneity in the endosperm. Using data from snRNA-Seq, the authors identified two distinct cell types in the chalazal domain based on gene expression analysis and cell cycle phase. Using the allelic gene expression data from their reciprocal crosses the authors found that there is almost no heterogeneity in the maternally expressed genes whereas paternally expressed genes show biased expression in the chalazal endosperm. Thus, this study identifies a cell-type and imprinting heterogeneity and provides a unique opportunity to compare gene expression, cell cycle, and imprinting in seemingly complex endosperm domains. Summary by (Vijaya Batthula @Vijaya_Batthula). bioRxiv. 10.1101/2020.08.25.267476.
Arabidopsis seeds consist of various tissue like seed coat, embryo, and endosperm. The endosperm provides the nutrient supplies to the growing embryo and has three domains namely, micropylar (surrounding embryo), chalazal (opposite end of the seed), and peripheral (in between micropylar and chalazal) domain. Even though there is transcriptomic data available for these different domains of endosperm, it is not yet known whether there is cell-type heterogeneity within these domains. In this paper Picard et al. carried out single nucleus RNA-sequencing (snRNA-Seq) on F1 seeds from a reciprocal cross between the Columbia (Col-0) and Cape Verde Islands (Cvi-0) to identify cell-type and imprinting heterogeneity in the endosperm. Using data from snRNA-Seq, the authors identified two distinct cell types in the chalazal domain based on gene expression analysis and cell cycle phase. Using the allelic gene expression data from their reciprocal crosses the authors found that there is almost no heterogeneity in the maternally expressed genes whereas paternally expressed genes show biased expression in the chalazal endosperm. Thus, this study identifies a cell-type and imprinting heterogeneity and provides a unique opportunity to compare gene expression, cell cycle, and imprinting in seemingly complex endosperm domains. Summary by (Vijaya Batthula @Vijaya_Batthula). bioRxiv. 10.1101/2020.08.25.267476.
The transcription factor bZIP60 modulates the heat shock response in maize
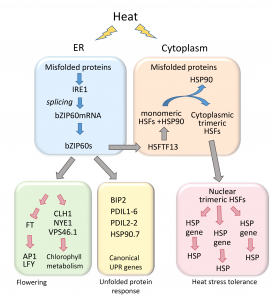 During heat stress, heat shock transcription factors upregulate heat shock protein genes and give rise to the heat shock response (HSR) that occurs in the cytoplasm, while in the endoplasmic reticulum (ER), the unfolded protein response (UPR) is also activated during heat stress. Both HSR and UPR cause the accumulation of misfolded proteins, a condition in the ER known as ER stress. Li et al. investigated the role of the Basic Leucine Zipper 60 (bZIP60) transcription factor, which plays an important role in UPR during heat stress and has previously been shown to be active following heat-induced splicing by an ER-membrane associated splicing factor. During elevated temperature, from 31ᵒC to 37ᵒC, the spliced form of bZIP60 and other HSR- and UPR-induced genes are gradually upregulated along with an increase in autophagy. bZIP60 activates the expression of HSFTF13, a type A heat shock factor, and this activation depends on the UPR-cis element in the promoter of this gene. The heat-induced “pale leaves” phenotype and senescence of the bzip60 maize mutant likely corresponds to the decreased expression of chlorophyll synthesis genes and increased expression of autophagy genes. Overall, bZIP60 mRNA could be a reliable biomarker for the UPR, and provides a link between HSR and UPR. (Summary by Min May Wong @wongminmay) Plant Cell 10.1105/tpc.20.00260
During heat stress, heat shock transcription factors upregulate heat shock protein genes and give rise to the heat shock response (HSR) that occurs in the cytoplasm, while in the endoplasmic reticulum (ER), the unfolded protein response (UPR) is also activated during heat stress. Both HSR and UPR cause the accumulation of misfolded proteins, a condition in the ER known as ER stress. Li et al. investigated the role of the Basic Leucine Zipper 60 (bZIP60) transcription factor, which plays an important role in UPR during heat stress and has previously been shown to be active following heat-induced splicing by an ER-membrane associated splicing factor. During elevated temperature, from 31ᵒC to 37ᵒC, the spliced form of bZIP60 and other HSR- and UPR-induced genes are gradually upregulated along with an increase in autophagy. bZIP60 activates the expression of HSFTF13, a type A heat shock factor, and this activation depends on the UPR-cis element in the promoter of this gene. The heat-induced “pale leaves” phenotype and senescence of the bzip60 maize mutant likely corresponds to the decreased expression of chlorophyll synthesis genes and increased expression of autophagy genes. Overall, bZIP60 mRNA could be a reliable biomarker for the UPR, and provides a link between HSR and UPR. (Summary by Min May Wong @wongminmay) Plant Cell 10.1105/tpc.20.00260
More than photosynthesis: The chloroplast’s role in plant cell defense pathways ($)
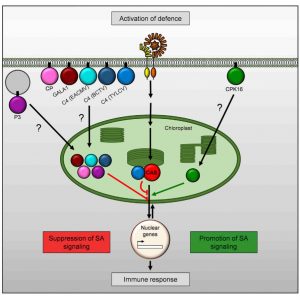 Chloroplasts are involved in various plant cell functions outside of photosynthesis including defense activation. How is the chloroplast able to do so? In this study, Medina-Puche et al. characterized the molecular function and cellular localization of the Tomato yellow leaf curl virus-derived C4 protein. The authors show that when expressed in plant cells, the protein initially localizes to the plasma membrane (PM), but re-localizes to the chloroplast when the cell initiates an immune response. This re-localization aids viral infection as the C4 protein can suppress chloroplast-specific defense mechanisms, specifically the biosynthesis of the defense hormone salicylic acid. The authors took inspiration from this mechanism, noting that the C4 protein contains both PM and chloroplast localization signals, and screened the Arabidopsis proteome for similar localization signatures to identify molecular players in chloroplast-mediated immunity. They identified the Calcium-Protein Kinase 16 (CAK16), which like the C4 protein initially localizes to the PM but upon detection of biotic threat re-localizes to the chloroplast and contributes positively towards chloroplast immune response. The authors thus conclude that “Based on the results presented here, we propose that a protein re-localization-dependent pathway physically linking PM and chloroplasts and regulating defense exists in plants, and that this pathway has been co-opted by pathogens during evolution to suppress defense responses and promote virulence.” (Summary by Benjamin Jin) Cell 10.1016/j.cell.2020.07.020
Chloroplasts are involved in various plant cell functions outside of photosynthesis including defense activation. How is the chloroplast able to do so? In this study, Medina-Puche et al. characterized the molecular function and cellular localization of the Tomato yellow leaf curl virus-derived C4 protein. The authors show that when expressed in plant cells, the protein initially localizes to the plasma membrane (PM), but re-localizes to the chloroplast when the cell initiates an immune response. This re-localization aids viral infection as the C4 protein can suppress chloroplast-specific defense mechanisms, specifically the biosynthesis of the defense hormone salicylic acid. The authors took inspiration from this mechanism, noting that the C4 protein contains both PM and chloroplast localization signals, and screened the Arabidopsis proteome for similar localization signatures to identify molecular players in chloroplast-mediated immunity. They identified the Calcium-Protein Kinase 16 (CAK16), which like the C4 protein initially localizes to the PM but upon detection of biotic threat re-localizes to the chloroplast and contributes positively towards chloroplast immune response. The authors thus conclude that “Based on the results presented here, we propose that a protein re-localization-dependent pathway physically linking PM and chloroplasts and regulating defense exists in plants, and that this pathway has been co-opted by pathogens during evolution to suppress defense responses and promote virulence.” (Summary by Benjamin Jin) Cell 10.1016/j.cell.2020.07.020
Root angle modifications by the DRO1 homolog improve rice yields in saline paddy fields
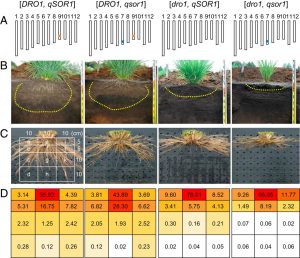 The spatial configuration of roots is a complex and critical trait for productivity that is impacted by abiotic stresses such as drought, waterlogging, and salinity. Soil salinity is expected to affect 50% of agricultural land worldwide by 2050. This research describes that the loss of function of qSOR1 (quantitative trait locus for SOIL SURFACE ROOTING 1) “qsor1-NIL” shows a shallow root growth angle (RGA) developing more on the soil surface. However, this QTL is a homolog of DRO1 (DEEPER ROOTING 1) which is a known regulator of RGA for deeper rooting. While a deep root system is beneficial to enhance drought avoidance, a shallow phenotype can facilitate the acquisition of phosphorus and facilitate growth in waterlogged areas. This response is negatively regulated via auxin signaling as qSOR1 expression declines rapidly after exogenous hormone application. Although DRO1 and qSOR1 show differential control of RGA, the C-terminal region among homologs is well conserved and determinant for proper function. Gravitropism sensing occurs in the root tips, and DRO1 is expressed in the whole root meristems except for the columella cells, whereas qSOR1 is specifically expressed in the columella cells. Saline conditions weaken qsor1-NIL rice root gravitropic responses similarly to those of wild-type roots when germinated in vitro. In contrast, in a field trial, qsor1-NIL rice yielded 15% more than the control in saline soil. Although the molecular mechanisms for the acquired increased yield need to be further investigated, this research highlights the potential of DRO1 homologs to improve salinity tolerance. (Summary by Camila Ribeiro @camicribeiro86) PNAS 10.1073/pnas.2005911117
The spatial configuration of roots is a complex and critical trait for productivity that is impacted by abiotic stresses such as drought, waterlogging, and salinity. Soil salinity is expected to affect 50% of agricultural land worldwide by 2050. This research describes that the loss of function of qSOR1 (quantitative trait locus for SOIL SURFACE ROOTING 1) “qsor1-NIL” shows a shallow root growth angle (RGA) developing more on the soil surface. However, this QTL is a homolog of DRO1 (DEEPER ROOTING 1) which is a known regulator of RGA for deeper rooting. While a deep root system is beneficial to enhance drought avoidance, a shallow phenotype can facilitate the acquisition of phosphorus and facilitate growth in waterlogged areas. This response is negatively regulated via auxin signaling as qSOR1 expression declines rapidly after exogenous hormone application. Although DRO1 and qSOR1 show differential control of RGA, the C-terminal region among homologs is well conserved and determinant for proper function. Gravitropism sensing occurs in the root tips, and DRO1 is expressed in the whole root meristems except for the columella cells, whereas qSOR1 is specifically expressed in the columella cells. Saline conditions weaken qsor1-NIL rice root gravitropic responses similarly to those of wild-type roots when germinated in vitro. In contrast, in a field trial, qsor1-NIL rice yielded 15% more than the control in saline soil. Although the molecular mechanisms for the acquired increased yield need to be further investigated, this research highlights the potential of DRO1 homologs to improve salinity tolerance. (Summary by Camila Ribeiro @camicribeiro86) PNAS 10.1073/pnas.2005911117
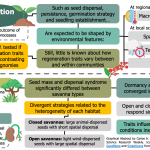
Do regeneration traits vary according to vegetation structure? A case study for savannas ($)
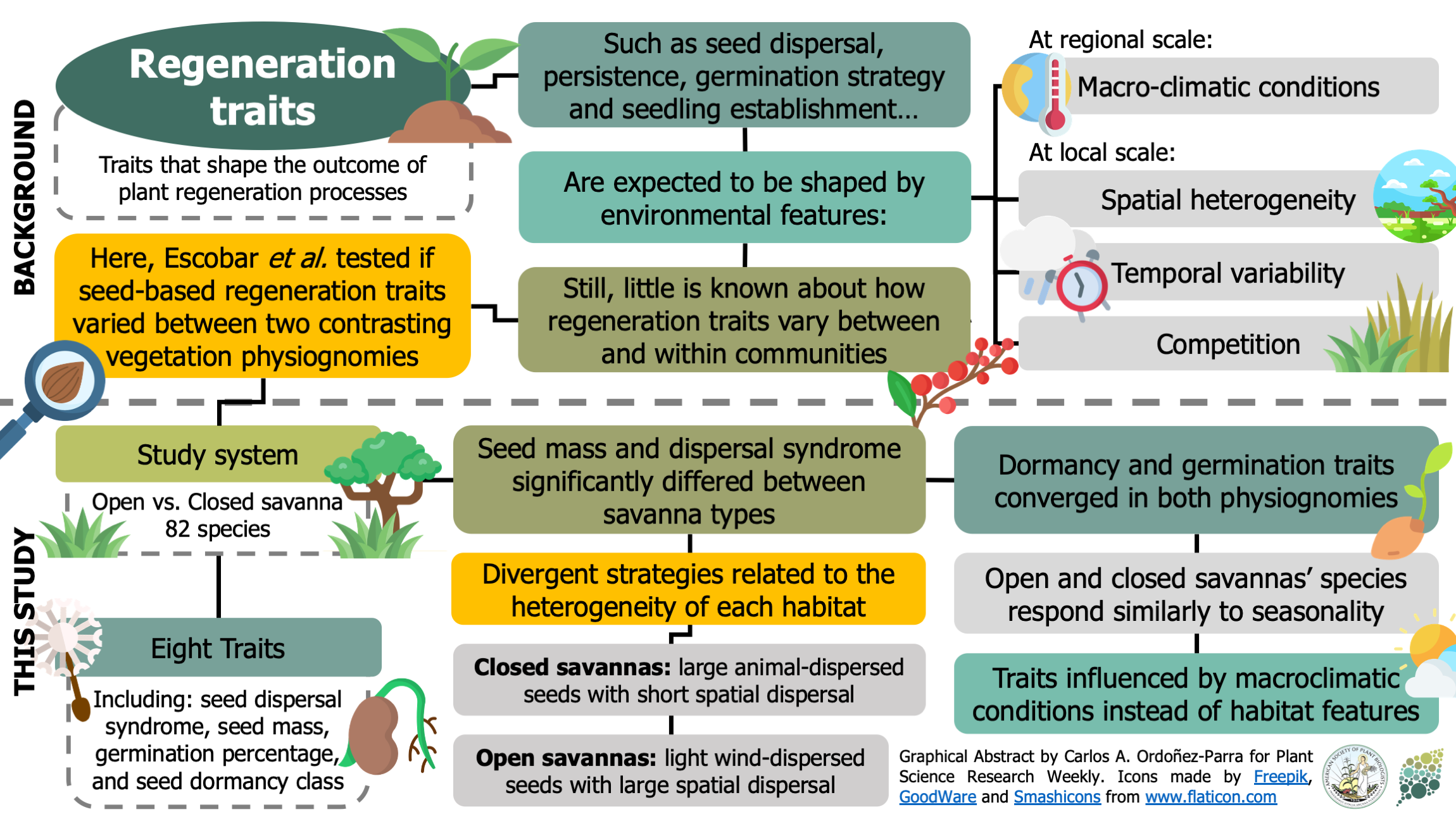 Regeneration traits –such as germination strategies, seed dormancy, and seedling establishment– are presumably shaped by environmental variables that operate at different temporal and spatial scales. However, empirical evidence about the variation is lacking for most regeneration seed traits. Here, Escobar et al. tested if seed-based regeneration traits varied between two vegetation habitats with contrasting environmental heterogeneity levels. Eight seed functional traits were assessed in 82 species from an open (i.e., higher heterogeneity) and a closed (i.e., lower heterogeneity) savanna in the Brazilian Cerrado. From these, only seed mass and dispersal syndrome significantly differed between habitats, with lighter and wind-dispersed seeds prevailing in open savannas. This divergence was attributed to the differences in each habitat’s environmental heterogeneity, suggesting that more variable habitats favor seeds with higher dispersal capacity. On the other hand, dormancy and germination traits were similar among savanna types, meaning these are controlled by the macroclimatic conditions rather than vegetation structure. Consequently, this study provides exciting insights into the effect of different environmental filters on seed-based regeneration strategies, helping our understanding of the rules behind plant community assembly. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) J. Veg. Sci. 10.1111/jvs.12940
Regeneration traits –such as germination strategies, seed dormancy, and seedling establishment– are presumably shaped by environmental variables that operate at different temporal and spatial scales. However, empirical evidence about the variation is lacking for most regeneration seed traits. Here, Escobar et al. tested if seed-based regeneration traits varied between two vegetation habitats with contrasting environmental heterogeneity levels. Eight seed functional traits were assessed in 82 species from an open (i.e., higher heterogeneity) and a closed (i.e., lower heterogeneity) savanna in the Brazilian Cerrado. From these, only seed mass and dispersal syndrome significantly differed between habitats, with lighter and wind-dispersed seeds prevailing in open savannas. This divergence was attributed to the differences in each habitat’s environmental heterogeneity, suggesting that more variable habitats favor seeds with higher dispersal capacity. On the other hand, dormancy and germination traits were similar among savanna types, meaning these are controlled by the macroclimatic conditions rather than vegetation structure. Consequently, this study provides exciting insights into the effect of different environmental filters on seed-based regeneration strategies, helping our understanding of the rules behind plant community assembly. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) J. Veg. Sci. 10.1111/jvs.12940