Plant Science Research Weekly: October 30
 This week’s Plant Science Research Weekly was guest-edited by Batthula Vijaya Lakshmi Vadde. She is currently a postdoc at Adrienne Roeder’s lab, Cornell University studying giant cell patterning in Arabidopsis sepals. Vijaya did her Ph.D. on trichome development in Arabidopsis with Utpal Nath from the Indian Institute of Science, Bangalore, India. Her specific research interests are developmental biology, genetics, cell biology, with a general interest in most of the scientific areas. She is an active member of Plantae mainly, contributing to the Plant Science Research Weekly, and you can find her Plantae profile at Vijaya Batthula and her summary contributions here. Apart from science, she is interested in photography (nature, long exposure, and star photography) and traveling. You can also find Vijaya on Twitter @Vijaya_batthula
This week’s Plant Science Research Weekly was guest-edited by Batthula Vijaya Lakshmi Vadde. She is currently a postdoc at Adrienne Roeder’s lab, Cornell University studying giant cell patterning in Arabidopsis sepals. Vijaya did her Ph.D. on trichome development in Arabidopsis with Utpal Nath from the Indian Institute of Science, Bangalore, India. Her specific research interests are developmental biology, genetics, cell biology, with a general interest in most of the scientific areas. She is an active member of Plantae mainly, contributing to the Plant Science Research Weekly, and you can find her Plantae profile at Vijaya Batthula and her summary contributions here. Apart from science, she is interested in photography (nature, long exposure, and star photography) and traveling. You can also find Vijaya on Twitter @Vijaya_batthula
Identification of a unique ZIP transporter involved in zinc uptake via the arbuscular mycorrhizal fungal pathway
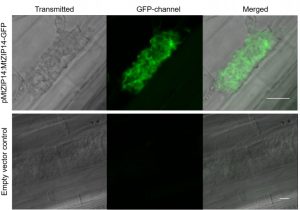 Last week, PSRW presented two review papers regarding host plant interactions with microbial populations, particularly for plant nutrient intake. Watt-Williams et al. utilize such knowledge for their paper, performing an RNA-seq dataset to identify a novel zinc transporter in Medicago truncatula. Zinc is a vital nutrient; its uptake in plants is commonly facilitated via associations with arbuscular mycorrhizal (AM) fungi. However, the plant’s molecular players in this fungi-plant-mediated zinc uptake are relatively uncharacterized. To untangle this complex process, the authors analyzed the transcriptomes of M. truncatula under varying levels of soil zinc concentrations, with or without AM association. One gene in particular, MtZIP14, was chosen for experimental analysis, as it was only expressed in AM-colonized root cells and had an expression pattern suggesting involvement in the zinc uptake pathway. MtZIP14 localizes to the peri-arbuscular membrane of AM-colonized cells, characteristic of a fungal-plant transporter, and demonstrates zinc transport ability when expressed in yeast. M. truncatula knock-down and knock-out mutants of MtZIP14 demonstrated altered expression of other ZIP genes, alongside decreased biomass, lower shoot zinc concentrations, and impaired AM colonization. These results open the gates towards better understanding how plants interact with their environment for their own well-being, information with sure agricultural and ecological applications. (Summary by Benjamin Jin) bioRxiv 10.1101/2020.09.28.317669
Last week, PSRW presented two review papers regarding host plant interactions with microbial populations, particularly for plant nutrient intake. Watt-Williams et al. utilize such knowledge for their paper, performing an RNA-seq dataset to identify a novel zinc transporter in Medicago truncatula. Zinc is a vital nutrient; its uptake in plants is commonly facilitated via associations with arbuscular mycorrhizal (AM) fungi. However, the plant’s molecular players in this fungi-plant-mediated zinc uptake are relatively uncharacterized. To untangle this complex process, the authors analyzed the transcriptomes of M. truncatula under varying levels of soil zinc concentrations, with or without AM association. One gene in particular, MtZIP14, was chosen for experimental analysis, as it was only expressed in AM-colonized root cells and had an expression pattern suggesting involvement in the zinc uptake pathway. MtZIP14 localizes to the peri-arbuscular membrane of AM-colonized cells, characteristic of a fungal-plant transporter, and demonstrates zinc transport ability when expressed in yeast. M. truncatula knock-down and knock-out mutants of MtZIP14 demonstrated altered expression of other ZIP genes, alongside decreased biomass, lower shoot zinc concentrations, and impaired AM colonization. These results open the gates towards better understanding how plants interact with their environment for their own well-being, information with sure agricultural and ecological applications. (Summary by Benjamin Jin) bioRxiv 10.1101/2020.09.28.317669
Transpiration from tomato fruit occurs primarily via trichome-associated transcuticular polar pores
 Post-harvest water loss is one of the major challenges in handling fresh agricultural products, yet its cause and mechanism are not well understood. In terrestrial plants, stomata are the primary regulators of water flux; but when the stomata are closed or in astomatous organs such as tomato fruits, the hydrophobic layer “cuticle” is the main barrier for transpiration. Using tomato cultivars with low and high fruit transpiration rates, Fich et al. show that the stem scar and cuticle contribute to the postharvest water loss in tomato fruits. As the stem scars heal by suberization a few days after the fruit harvest, the cuticle maintains the main source of transpiration. However, despite the considerable variation throughout the cultivars, neither cuticular cutin nor wax total abundance and composition explained the transpiration rate of the tomato fruits. Instead, staining by toluidine blue, they found that the hydrophilic dye enters the fruit through small hydrophilic permeable pores corresponding to the broken trichome bases. The pores that let the dye in, let the water out and their density strongly correlate with the fruits transpiration rates. Understanding this novel mechanism provides a base for agricultural practices to select cultivars with more resistance to postharvest water loss. (Summary by Roxana Khoshravesh @RoxiKh) Plant Physiol. 10.1104/pp.20.01105
Post-harvest water loss is one of the major challenges in handling fresh agricultural products, yet its cause and mechanism are not well understood. In terrestrial plants, stomata are the primary regulators of water flux; but when the stomata are closed or in astomatous organs such as tomato fruits, the hydrophobic layer “cuticle” is the main barrier for transpiration. Using tomato cultivars with low and high fruit transpiration rates, Fich et al. show that the stem scar and cuticle contribute to the postharvest water loss in tomato fruits. As the stem scars heal by suberization a few days after the fruit harvest, the cuticle maintains the main source of transpiration. However, despite the considerable variation throughout the cultivars, neither cuticular cutin nor wax total abundance and composition explained the transpiration rate of the tomato fruits. Instead, staining by toluidine blue, they found that the hydrophilic dye enters the fruit through small hydrophilic permeable pores corresponding to the broken trichome bases. The pores that let the dye in, let the water out and their density strongly correlate with the fruits transpiration rates. Understanding this novel mechanism provides a base for agricultural practices to select cultivars with more resistance to postharvest water loss. (Summary by Roxana Khoshravesh @RoxiKh) Plant Physiol. 10.1104/pp.20.01105
The structure of a triple complex of plant photosystem I with ferredoxin and plastocyanin
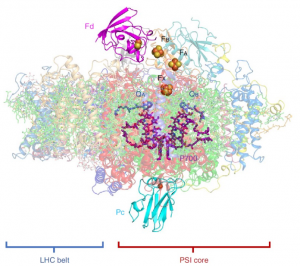 Photosystem I (PSI) is a membrane-bound protein complex that plays a central role in the conversion of solar energy to chemical energy to drive assimilate production. Photosystem I is an enzyme complex that transfers electrons from plastocyanin (Pc) to ferredoxin (Fd). Here Caspy et al. have characterized a high-resolution ultrastructure of the entire complex, PSI associated with Pc and Fd. The authors added purified Pc and Fd to pure Pisum sativum PSI crystals obtained previously. They used cryo-EM to arrive at a high-resolution structure with all the details of binding sites and interactions within it. The study established that the transient binding of Fd to PSI is essential for its quick reduction by PSI to enable a high turnover of the reaction. In the PSI–plastocyanin binding site, the reduced electron carriers are in the lumen side of the PSI and transfer is mediated by cytochrome c6 and plastocyanin. It was found that Fd relies on electrostatic interactions from binding proteins PsaA, PsaC and PsaE whereas plastocyanin depends on hydrophobic interactions with PsaA and PsaB. (Summary by Arun K. Shanker @arunshanker) Nature Plants 10.1038/s41477-020-00779-9
Photosystem I (PSI) is a membrane-bound protein complex that plays a central role in the conversion of solar energy to chemical energy to drive assimilate production. Photosystem I is an enzyme complex that transfers electrons from plastocyanin (Pc) to ferredoxin (Fd). Here Caspy et al. have characterized a high-resolution ultrastructure of the entire complex, PSI associated with Pc and Fd. The authors added purified Pc and Fd to pure Pisum sativum PSI crystals obtained previously. They used cryo-EM to arrive at a high-resolution structure with all the details of binding sites and interactions within it. The study established that the transient binding of Fd to PSI is essential for its quick reduction by PSI to enable a high turnover of the reaction. In the PSI–plastocyanin binding site, the reduced electron carriers are in the lumen side of the PSI and transfer is mediated by cytochrome c6 and plastocyanin. It was found that Fd relies on electrostatic interactions from binding proteins PsaA, PsaC and PsaE whereas plastocyanin depends on hydrophobic interactions with PsaA and PsaB. (Summary by Arun K. Shanker @arunshanker) Nature Plants 10.1038/s41477-020-00779-9
TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T
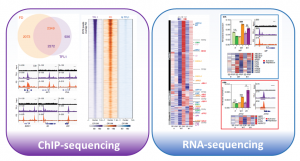 The onset of reproductive development is tightly regulated in response to environmental cues as early and delayed flowering greatly affects seed production. In Arabidopsis, the timing of flower formation is controlled by members of the Phosphatidyl Ethanolamine-Binding Protein (PEBP) family: TERMINAL FLOWER 1 (TFL1) promotes inflorescence fate whereas FLOWERING LOCUS T (FT) promotes floral fate. As PEBPs lack DNA binding activity, their interaction with FLOWERING LOCUS D (FD), a bZIP transcription factor in the apex is required to coordinate the expression of downstream genes. In this paper, Zhu and coworkers elegantly resolved the mechanisms underlying the antagonism between FT and TFL1 in shaping inflorescence architecture. Using the ChIP-sequencing experiment, the authors identified an overlap between TFL1 and FD-peaks, and reduced TFL1 chromatin occupancy in fd-1 mutant, indicating that FD is required to recruit TFL1 to target loci. Among the target genes, TFL1 and FT acts antagonistically on LEAFY (LFY), a floral meristem identity gene. Interestingly, three bZIP-binding sites in the second exon of LFY defined its spatio-temporal expression, as these cis-regulatory elements are necessary for repression via TFL1–FD in the shoot apex as well as direct activation via FT-FD in floral primordia. RNA-sequencing on wild-type, ft and, tfl1 mutant plants identified changes in transcriptional programs related to reproductive development and response to endogenous and abiotic signals. To conclude, plants modulate inflorescence architecture in response to seasonal changes by altering the balance between antagonistic PEBPs. At the molecular level, FT and TFL1 compete for bZIP binding sites in regulatory sequences of target genes, which regulate flower development and signaling pathways. (Summary by Michela Osnato @michela_osnato) Nat. Commun. https://doi.org/10.1038/s41467-020-18782-1
The onset of reproductive development is tightly regulated in response to environmental cues as early and delayed flowering greatly affects seed production. In Arabidopsis, the timing of flower formation is controlled by members of the Phosphatidyl Ethanolamine-Binding Protein (PEBP) family: TERMINAL FLOWER 1 (TFL1) promotes inflorescence fate whereas FLOWERING LOCUS T (FT) promotes floral fate. As PEBPs lack DNA binding activity, their interaction with FLOWERING LOCUS D (FD), a bZIP transcription factor in the apex is required to coordinate the expression of downstream genes. In this paper, Zhu and coworkers elegantly resolved the mechanisms underlying the antagonism between FT and TFL1 in shaping inflorescence architecture. Using the ChIP-sequencing experiment, the authors identified an overlap between TFL1 and FD-peaks, and reduced TFL1 chromatin occupancy in fd-1 mutant, indicating that FD is required to recruit TFL1 to target loci. Among the target genes, TFL1 and FT acts antagonistically on LEAFY (LFY), a floral meristem identity gene. Interestingly, three bZIP-binding sites in the second exon of LFY defined its spatio-temporal expression, as these cis-regulatory elements are necessary for repression via TFL1–FD in the shoot apex as well as direct activation via FT-FD in floral primordia. RNA-sequencing on wild-type, ft and, tfl1 mutant plants identified changes in transcriptional programs related to reproductive development and response to endogenous and abiotic signals. To conclude, plants modulate inflorescence architecture in response to seasonal changes by altering the balance between antagonistic PEBPs. At the molecular level, FT and TFL1 compete for bZIP binding sites in regulatory sequences of target genes, which regulate flower development and signaling pathways. (Summary by Michela Osnato @michela_osnato) Nat. Commun. https://doi.org/10.1038/s41467-020-18782-1
Coleorhiza‐enforced seed dormancy: a novel mechanism to control germination in grasses
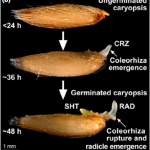
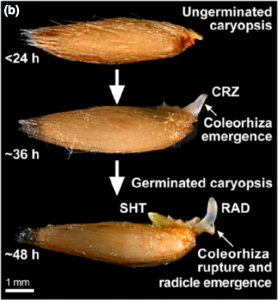 The germination process is completed when the radicle emerges from the seed. In eudicots, radicle growth is mechanically limited by the micropylar endosperm. As dormancy is broken and imbibition occurs, this structure is weakened so that radicle can pierce it. In grasses, the radicle is not surrounded by endosperm, but a sheath-like tissue called the coleorhiza. Still, little is known about the role of this structure in the regulation of radicle emergence. In this paper, Holloway and colleagues show that coleorhiza shapes radicle emergence in Avena fatua and that this control depends on the seed dormancy state. Dormant seeds could germinate if their coleorhiza were experimentally removed, confirming it constrains radicle’s emergence. Moreover, the authors show that if seeds are dormant upon imbibition, the coleorhiza becomes more robust and harder to pierce by the radicle. Contrastingly, if dormancy has been broken, the coleorhiza is weakened as imbibition occurs, facilitating radicle protrusion. These divergent behaviors arise from the recruitment of different groups of xyloglucan endo-transglycosylases/hydrolases that contribute to either loosen or harden coleorhiza cell walls, depending on seed dormancy state. As a result, this research provides exciting insights into the coleorhiza’s role in seed dormancy and the biochemical mechanisms behind it. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) New Phytol. 10.1111/nph.16948
The germination process is completed when the radicle emerges from the seed. In eudicots, radicle growth is mechanically limited by the micropylar endosperm. As dormancy is broken and imbibition occurs, this structure is weakened so that radicle can pierce it. In grasses, the radicle is not surrounded by endosperm, but a sheath-like tissue called the coleorhiza. Still, little is known about the role of this structure in the regulation of radicle emergence. In this paper, Holloway and colleagues show that coleorhiza shapes radicle emergence in Avena fatua and that this control depends on the seed dormancy state. Dormant seeds could germinate if their coleorhiza were experimentally removed, confirming it constrains radicle’s emergence. Moreover, the authors show that if seeds are dormant upon imbibition, the coleorhiza becomes more robust and harder to pierce by the radicle. Contrastingly, if dormancy has been broken, the coleorhiza is weakened as imbibition occurs, facilitating radicle protrusion. These divergent behaviors arise from the recruitment of different groups of xyloglucan endo-transglycosylases/hydrolases that contribute to either loosen or harden coleorhiza cell walls, depending on seed dormancy state. As a result, this research provides exciting insights into the coleorhiza’s role in seed dormancy and the biochemical mechanisms behind it. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) New Phytol. 10.1111/nph.16948