Plant Science Research Weekly: October 16, 2020
Review: The second decade of synthetic biology: 2010–2020
I guess if this were a normal year, we’d be spending time looking back over the past decade, but Covid-19 has made anything pre-2019 seem like a different lifetime. Still, here’s a retrospective you don’t want to miss. Meng and Ellis take us on a short walk down memory lane through the “second decade” of synthetic biology – and what a decade it’s been. I won’t give away too much here, because you should read this yourself, but we’ve come a long way since 2010. (The authors crowdsourced some of these ideas on Twitter, and acknowledge these conversations https://twitter.com/ProfTomEllis/status/1280982288797446144 – it’s interesting to ponder how Twitter might have contributed to SynBio over the past decade). What’s even more exciting is that later this year there will be a companion “look ahead” to 2030. I wonder where SynBio will take us by 2030? (Summary by Mary Williams @PlantTeaching) Nature Comms. 10.1038/s41467-020-19092-2
Review: Gemma cup and gemma development in Marchantia polymorpha
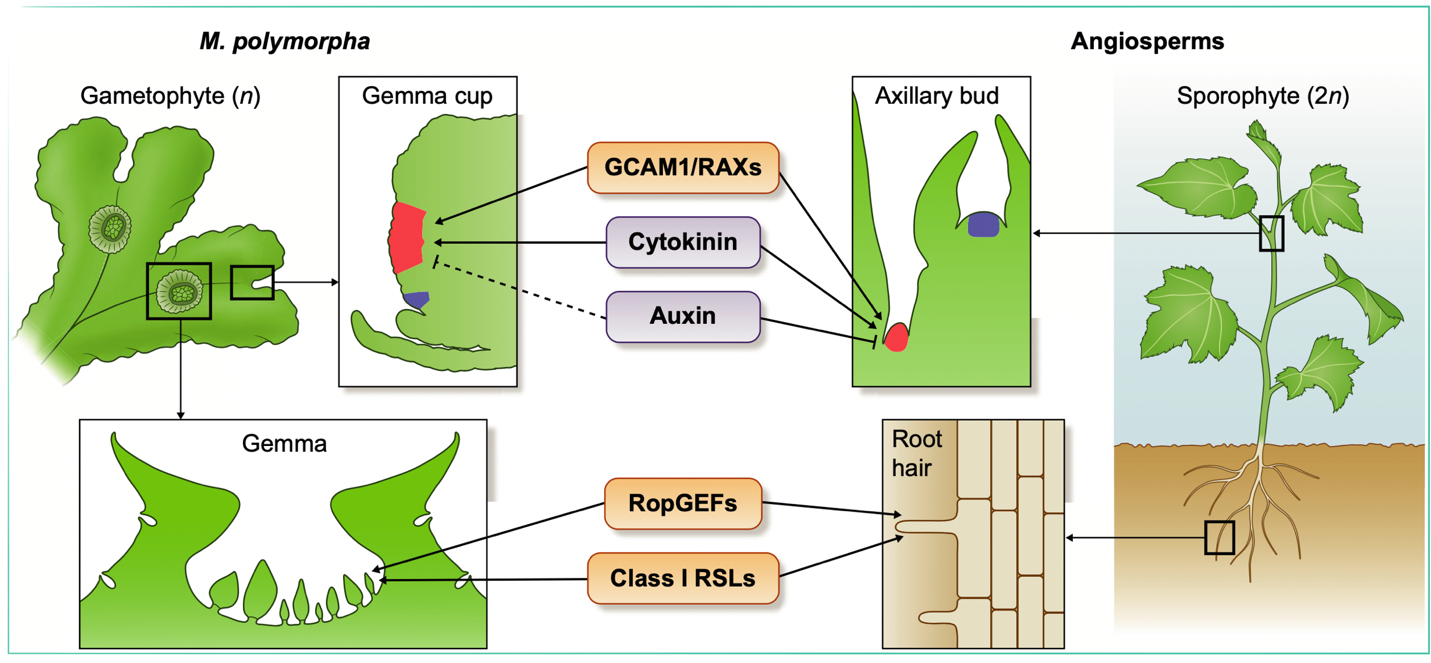
Plants have the astounding capacity to generate new plants derived from specialized organs (i.e., roots, leaves, and stems). This capacity, known as vegetative propagation, is used by horticulturists and farmers to propagate clonal varieties. This form of reproduction is fueled by the ability to regenerate and proliferate meristems from differentiated tissue. The liverwort model Marchantia polymorpha is a suitable candidate in which to study natural vegetative propagation given its low genetic redundancy and its capacity to reproduce asexually through gemmae, which are clonal propagules that contain two apical meristems and that develop inside specialized cup-shaped receptacles (gemmae cups). A recent review by Kato et al. summarizes known regulators involved in gemmae and gemmae cup development, and specially focuses on shared regulatory elements in Marchantia and angiosperms that control the formation of meristems from differentiated tissues. Examples include GEMMA CUP-ASSOCIATED MYB1 (GCAM1) and ROOT-HAIR DEFECTIVE SIX-LIKE (RSL1), transcription factors required for gemmae cup initiation and gemmae development, respectively. Both are conserved in angiosperms and regulate analogous processes; examples include the GCAM1 homologues Arabidopsis REGULATOR OF AXILLARY MERISTEM and Solanum lycopersicum Blind, and the RSLs, which are key regulators of root hair development in angiosperms and rhizoids in Physcomitrella patens. This review highlights that although gemmae and gemmae cups are specific to some Marchantia species, their development is controlled by regulatory modules present in other plant lineages. Thus, research into what controls gemmae and gemmae cup development could reveal central regulatory modules underlying organogenesis in land plants. (Summary by Jesus Leon @jesussaur) New Phytol. 10.1111/nph.16655
The love potion made by soil-borne fungi
 The basic requirements for plant growth are water, nutrients, and light. There are many other factors contributing to plant growth, including the interactions between plants and soil microorganisms. Soil microorganisms produce a large array of volatiles that can affect root architecture (e.g., some volatiles result in shortened primary roots and increased lateral root formation), but it remains unknown whether the volatiles affect directional root growth. To address this question, Moisan et al. designed a Y-tube olfactometer to test the directional preference of Brassica rapa primary root growth, with or without the presence of fungi-released volatiles. Of the four soil-borne fungi tested, only Rhizoctonia solani mediated directional root growth preference: B. rapa grew towards volatiles released by R. solani. Despite the primary root directional preference, the primary root length and total root dry weight were not affected by the tested fungal volatiles. Comparing volatile profiles between the four tested fungi revealed six volatiles that were unique to R. solani, none of which had been found to promote root growth previously; perhaps timing, duration, and concentration of volatiles as well as interplay between multiple volatiles contributed the disparity. This paper provides an expandable experimental setup for quantitatively investigating directional root growth. Interestingly, among the four tested fungi, R. solani was the only one pathogenic to B. rapa, suggesting volatiles can be a means by which a pathogenic fungus can manipulate plant growth, or as the authors put it – “fatal attraction.” (Summary by Yun-Ting Kao @YTingKao) Plant Cell Environ. 10.1111/pce.13890
The basic requirements for plant growth are water, nutrients, and light. There are many other factors contributing to plant growth, including the interactions between plants and soil microorganisms. Soil microorganisms produce a large array of volatiles that can affect root architecture (e.g., some volatiles result in shortened primary roots and increased lateral root formation), but it remains unknown whether the volatiles affect directional root growth. To address this question, Moisan et al. designed a Y-tube olfactometer to test the directional preference of Brassica rapa primary root growth, with or without the presence of fungi-released volatiles. Of the four soil-borne fungi tested, only Rhizoctonia solani mediated directional root growth preference: B. rapa grew towards volatiles released by R. solani. Despite the primary root directional preference, the primary root length and total root dry weight were not affected by the tested fungal volatiles. Comparing volatile profiles between the four tested fungi revealed six volatiles that were unique to R. solani, none of which had been found to promote root growth previously; perhaps timing, duration, and concentration of volatiles as well as interplay between multiple volatiles contributed the disparity. This paper provides an expandable experimental setup for quantitatively investigating directional root growth. Interestingly, among the four tested fungi, R. solani was the only one pathogenic to B. rapa, suggesting volatiles can be a means by which a pathogenic fungus can manipulate plant growth, or as the authors put it – “fatal attraction.” (Summary by Yun-Ting Kao @YTingKao) Plant Cell Environ. 10.1111/pce.13890
Multi-omics analyses of wild tomato introgression lines reveal a set of gene expression-metabolite-pathogen sensitivity interactions
 Domesticated tomato has been subjected to human selection to satisfy marketing and economic desires, leading to a loss of genetic diversity and elimination of fruits traits such as flavor, aroma and pathogen resistance. Introgression lines (ILs) that capture the genome of wild desert-adapted tomato Solanum pennellii in the background of cultivated tomato, S. lycopersicum, provide a remarkable tool to improve the resolution of Quantitative Trait Locus (QTL) mapping. Szymański and colleagues carried out transcriptome-metabolome-pathogen sensitivity analysis in breaker (slightly pink) and red ripe stages of 580 backcrossed inbred lines (BILs) and ILs. The multi-omics analysis identified genomic loci associated with hundreds of transcripts and metabolites related to tomato fruits development and ripening. The analysis uncovered core enzyme components of the tomato alkaloid pathway as well as a set of genes and metabolites promoting fungal resistance, which highlight the role of phenylpropanoids in fruit defenses. This large dataset will be an resource finding for molecular breeding in selection for flavor and pathogen resistance and is accessible through an online data browser https://szymanskilab.shinyapps.io/kilbil/. (Summary by Min May Wong @wongminmay) Nat. Genet. 10.1038/s41588-020-0690-6
Domesticated tomato has been subjected to human selection to satisfy marketing and economic desires, leading to a loss of genetic diversity and elimination of fruits traits such as flavor, aroma and pathogen resistance. Introgression lines (ILs) that capture the genome of wild desert-adapted tomato Solanum pennellii in the background of cultivated tomato, S. lycopersicum, provide a remarkable tool to improve the resolution of Quantitative Trait Locus (QTL) mapping. Szymański and colleagues carried out transcriptome-metabolome-pathogen sensitivity analysis in breaker (slightly pink) and red ripe stages of 580 backcrossed inbred lines (BILs) and ILs. The multi-omics analysis identified genomic loci associated with hundreds of transcripts and metabolites related to tomato fruits development and ripening. The analysis uncovered core enzyme components of the tomato alkaloid pathway as well as a set of genes and metabolites promoting fungal resistance, which highlight the role of phenylpropanoids in fruit defenses. This large dataset will be an resource finding for molecular breeding in selection for flavor and pathogen resistance and is accessible through an online data browser https://szymanskilab.shinyapps.io/kilbil/. (Summary by Min May Wong @wongminmay) Nat. Genet. 10.1038/s41588-020-0690-6
Active DNA demethylation controls defense gene regulation and antibacterial resistance
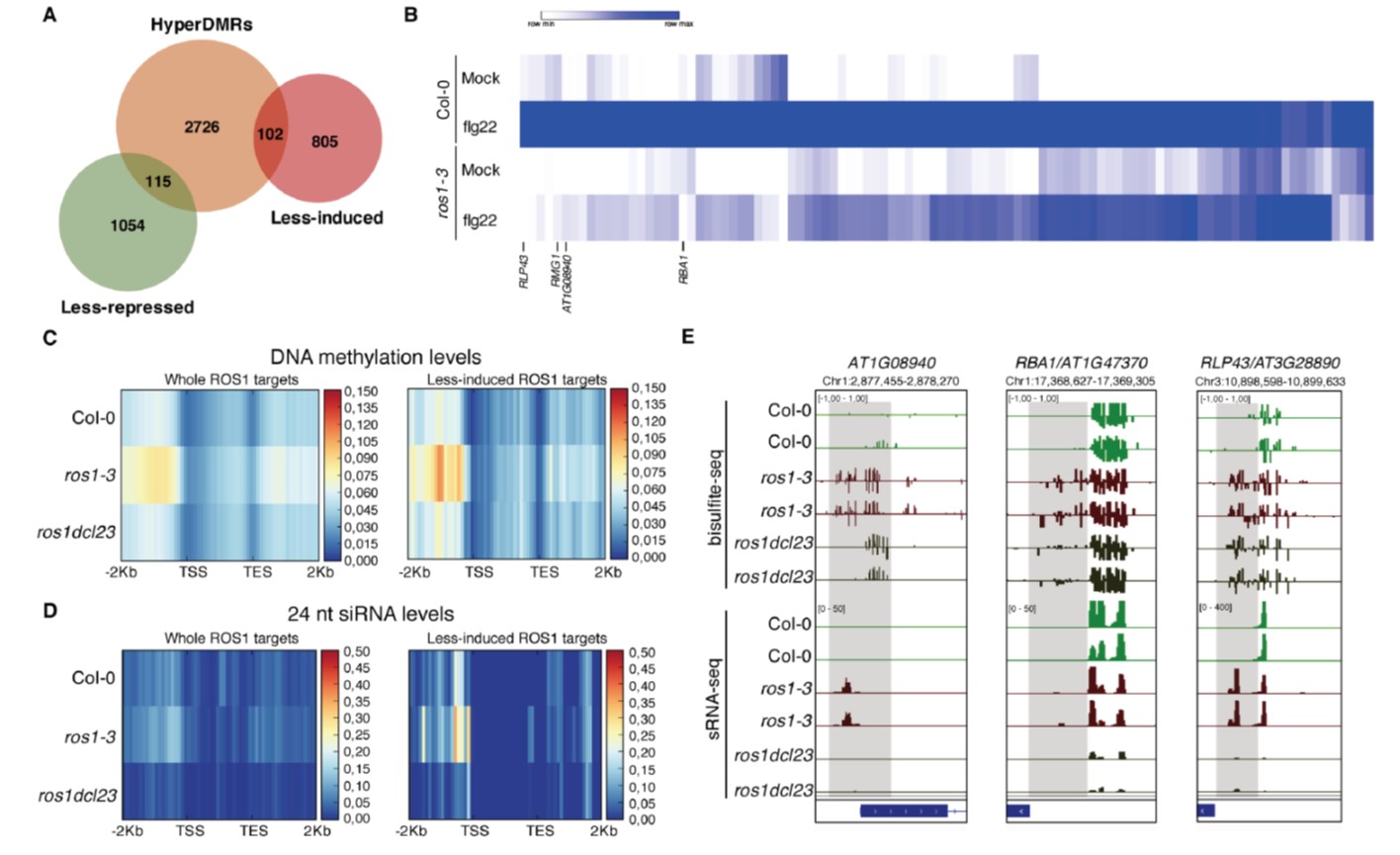
Epigenetic reprogramming, such as DNA methylation changes, has emerged as a crucial regulator of plant defense responses. Homeostasis of DNA methylation is controlled by a balance between methylation and demethylation. ROS1 (Repressor Of Silencing 1) is an Arabidopsis demethylases that is known to be a positive regulator of defense, but little is known about the mechanisms involved in this process. Halter et al. showed that ROS1 positively regulates plant defense against bacterial pathogens by antagonizing the function of DCL2/3 (DICER-LIKE 2/3), which play key roles in RNA-mediated DNA methylation (RdDM). A genome-wide analysis identified >2000 genes differentially regulated by ROS1 upon treatment of the immune elicitor flg22, among which 10% are demethylated by ROS1. Among such demethylated regions were WRKY transcription factor (TF) binding sites. The authors showed that hypermethylation at the promoter region of RLP43 (Receptor-Like Protein 43), a flg22 inducible gene, blocks binding of WRKY TFs to this DNA region. Furthermore, they showed that DNA demethylation in the promoter regions of defense-related genes, including RLP43, is important for the induction of these genes upon flg22 treatment and resistance against the bacterial pathogen Pseudomonas syringae. Taken together, this study reveals the molecular mechanisms underlying how ROS1-mediated demethylation plays critical roles in regulating plant defense responses and antibacterial resistance. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2020.09.30.321257
A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants
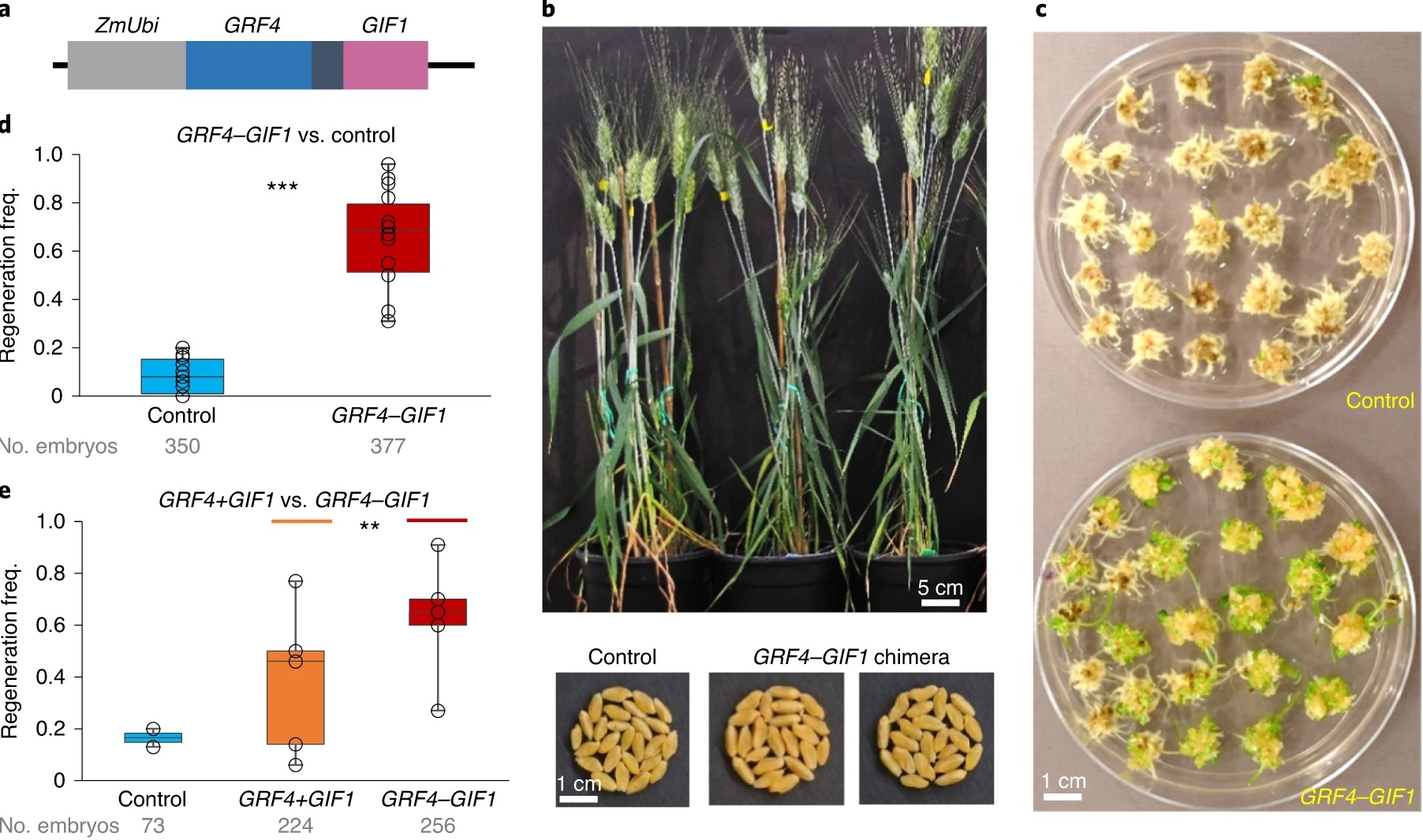
Producing a genetically-modified or -edited plant requires two distinct processes: DNA modification, followed by regeneration of a plant from the edited cells. The first process has been greatly enhanced by CRISPR/Cas9, but the second has continued to present challenges. Here, Debernardi et al. demonstrate a strategy to increase the rate and efficiency of plant regeneration. Previously, a small family of plant-specific transcription factors known as growth regulating factors (GRFs) were found to promote growth of leaves and other organs. The activity of GRFs is regulated in part by interaction with co-transcriptional regulators known as GRF-interacting factors (GIFs), and the GRFs and GIFs function as a complex. Debernardi et al. designed a chimeric GRF/GIF fusion protein which they showed accelerates regeneration of green shoots from callus tissue and can do so without culturing on cytokinins. Interestingly, when GRF and GIF were introduced as two independent genes on a single construct, the positive benefit was greatly reduced as compared to the expression of the single chimeric gene. The authors tested this strategy using several types of wheat and rice as well as citrus with generally positive outcomes. The authors observe that it might be possible to get even higher regeneration efficiencies by combining GRF-GIF with the meristem-promoting genes BBM and WUS2 that function earlier in differentiation and have been used previously to enhance regeneration. (Summary by Mary Williams @PlantTeaching) Nature Biotechnol. 10.1038/s41587-020-0703-0
Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9
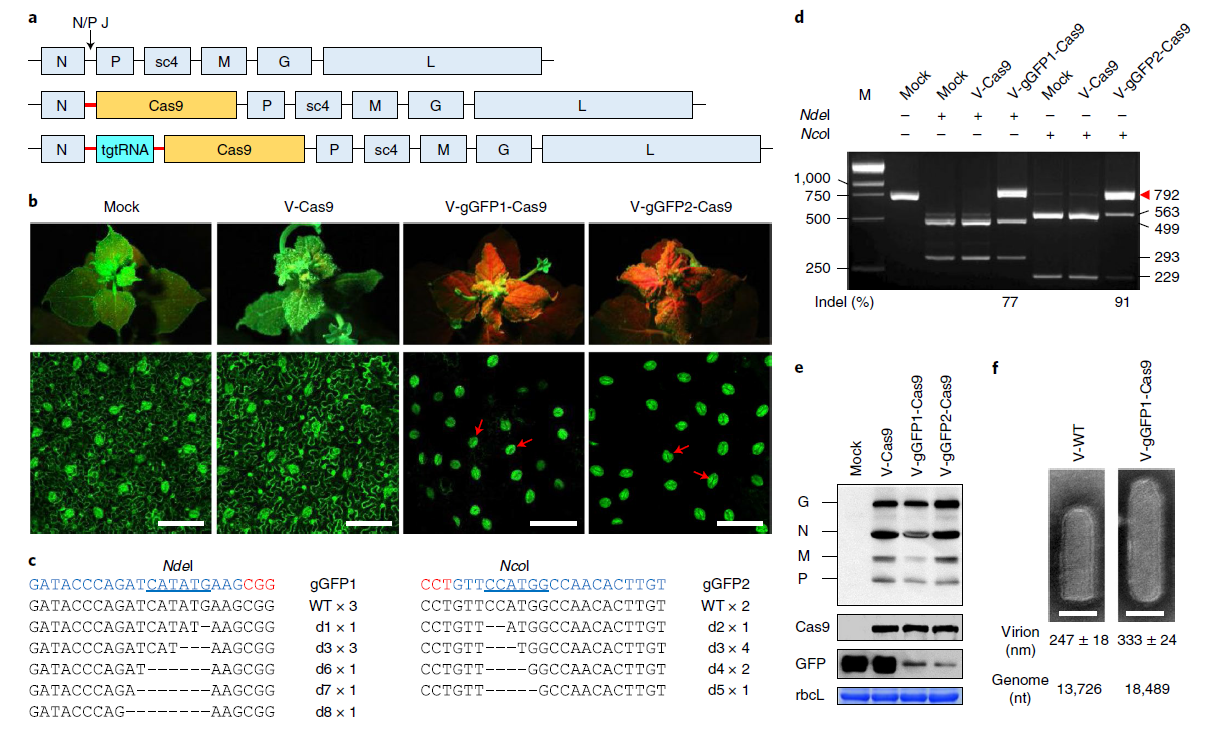
CRISPR-Cas is a revolutionary technology that has taken the science of genetic manipulation to higher levels by leaps and bounds. In spite of the ease with which it is used for genome editing, the present delivery methods are not without shortcomings; undesirable effects on non-target regions and possible damage to the genome must be addressed. Direct delivery of genetic information that is free of DNA is one way to overcome many of these shortcomings. Rhabdoviruses are negative strand RNA viruses (-ssRNA viruses) that have shown promise for mammalian genome editing. Here the authors inserted Streptococcus pyogenes Cas9 and gRNA sequences into the sonchus yellow net rhabdovirus genome, and the infected plants contained the mutations that were specifically targeted. The effect on parts of the genome that were not targeted was minimal. One of the possible limitations of this technique is the host range specificity of the virus. (Summary by Arun K. Shanker @arunshanker) Nature Plants http://dx.doi.org/10.1038/s41477-020-0704-5
Burning grasses, poor seeds: Post-fire reproduction of early-flowering Neotropical savanna grasses produces low-quality seeds ($)
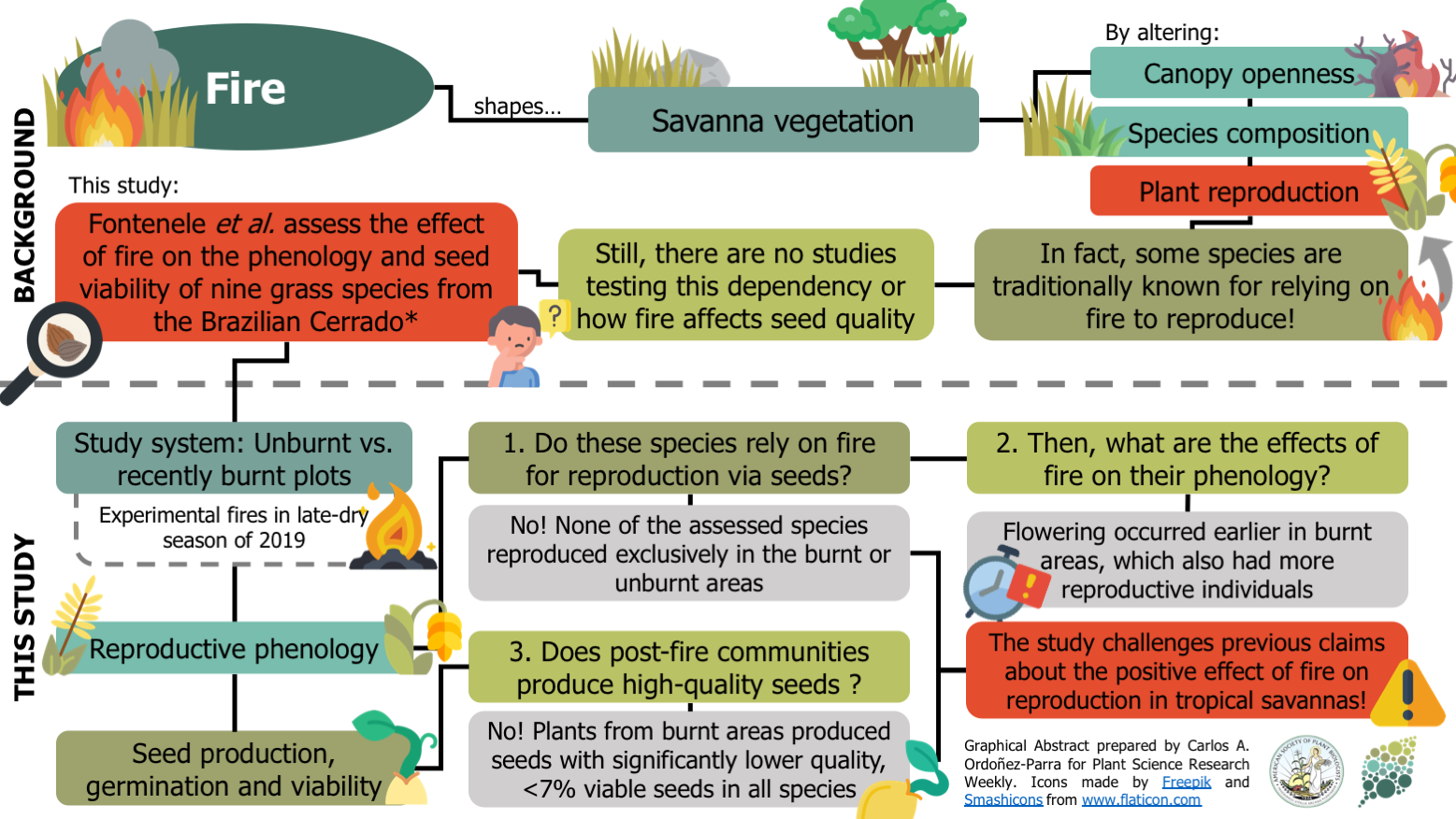
Fire is a disturbance that underpins several ecological processes in tropical savannas. For instance, some plant species from these ecosystems are traditionally known for relying on fires to reproduce. However, this dependency remains to be formally tested. Also, the impact of fire on seed quality has not been assessed, limiting our understanding of plant reproductive success after wildfires. Here, Fontenele and colleagues evaluate the effects of fire on the reproductive phenology and seed viability of nine grass species from the Brazilian Cerrado. Although none of the species reproduced exclusively in either burn or unburnt sites, areas submitted to experimental fires had more reproductive individuals and flowered sooner. As a result, the authors discuss that fire is not totally necessary for plant reproduction but can stimulate it. Still, grasses from burnt areas produced seed sets with significantly lower quality, with none of them making more than 7% of viable seeds. Given this, the study challenges previous claims about the positive relationship between fire and plant reproduction in Neotropical savannas. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Plant Ecol. 10.1007/s11258-020-01080-7
Seed germination of mudflat species responds differently to prior exposure to hypoxic (flooded) environments ($)
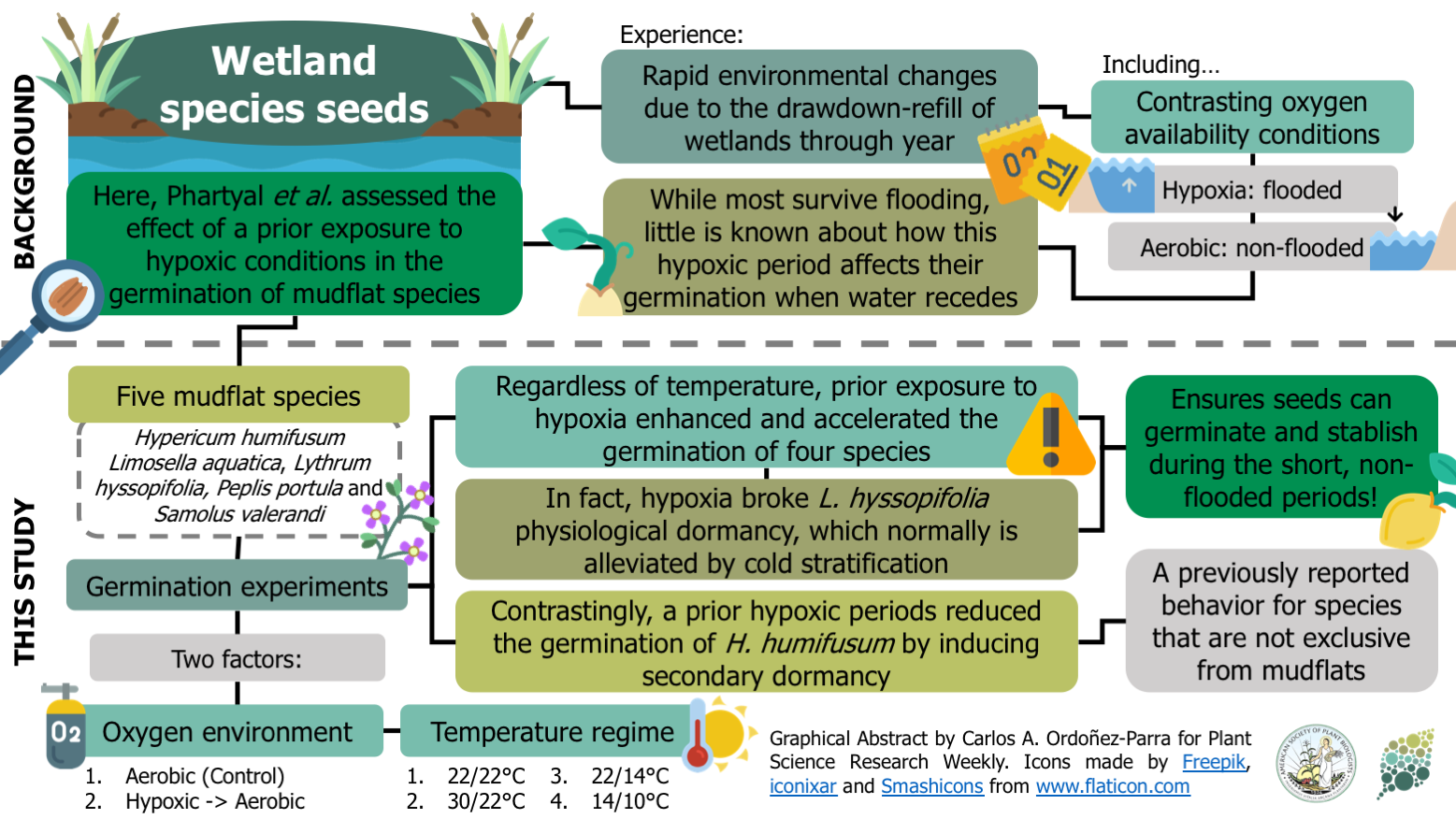
Mudflats species experience rapid changes in their environment due to wetlands’ drawdown and refill throughout the year. For instance, species that inhabit these sites deal with contrasting oxygen environments: hypoxic when the water level is high, and aerobic when it recedes. Most studies have aimed to examine the effect of hypoxia on seed viability. Still, little is known about how these conditions could affect seed germination when oxygen becomes available again. Here, Phartyal and colleagues assess the effect of prior exposure to hypoxic conditions on the germination of five mudflat species from Central Europe. The germination of four of the assessed species increased when seeds experienced hypoxia. For instance, hypoxic conditions seemed enough to alleviate the physiological dormancy of Lythrum hyssopifolia, which generally requires cold stratification to germinate. Given this, hypoxia’s stimulation of germination could be seen as a strategy to maximize growth when the water recedes, usually for a very short time. In contrast, the germination of Hypericum humifusum, which inhabits environments that rarely experience flooding, was reduced when exposed to hypoxia, implying that flooding induces secondary dormancy in this species. Consequently, this study provides insights into the importance of germination responses in the assembly of wetland plant communities. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Seed Sci. Res. 10.1017/S0960258520000240