Plant Science Research Weekly: November 8
Review: Exchange of small regulatory RNAs between plants and their pests
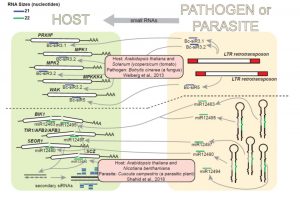 Trans-species small RNAs are the latest class in the family of signals that move between plants and their attackers. Hudzik et al. review this topic, covering small RNAs that move from plant to pest and from pest to plant. The transmitted RNAs function by interfering with gene expression in the recipient. Naturally-occurring plant-produced small RNAs have been identified that target fungal and oomycete pathogens. Recently, plant-produced extracellular vesicles (EVs) have been identified carrying defense proteins, small RNAs, and a new class called tiny RNAs (tyRNAs). Transgenic plants have also been created that produce small RNAs targeting nematodes, insects, and parasitic plants, as well as fungi and oomycites, in a process called host-induced gene silencing (HIGS). Conversely, many of these pests and pathogens produce small RNAs that target plant genes. The authors also describe the roles of small RNAs in the interactions between parastic plants and their hosts, and speculate on the possible evolution of trans-species RNAs. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.00931
Trans-species small RNAs are the latest class in the family of signals that move between plants and their attackers. Hudzik et al. review this topic, covering small RNAs that move from plant to pest and from pest to plant. The transmitted RNAs function by interfering with gene expression in the recipient. Naturally-occurring plant-produced small RNAs have been identified that target fungal and oomycete pathogens. Recently, plant-produced extracellular vesicles (EVs) have been identified carrying defense proteins, small RNAs, and a new class called tiny RNAs (tyRNAs). Transgenic plants have also been created that produce small RNAs targeting nematodes, insects, and parasitic plants, as well as fungi and oomycites, in a process called host-induced gene silencing (HIGS). Conversely, many of these pests and pathogens produce small RNAs that target plant genes. The authors also describe the roles of small RNAs in the interactions between parastic plants and their hosts, and speculate on the possible evolution of trans-species RNAs. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.00931
Review: Mosses in biotechnology
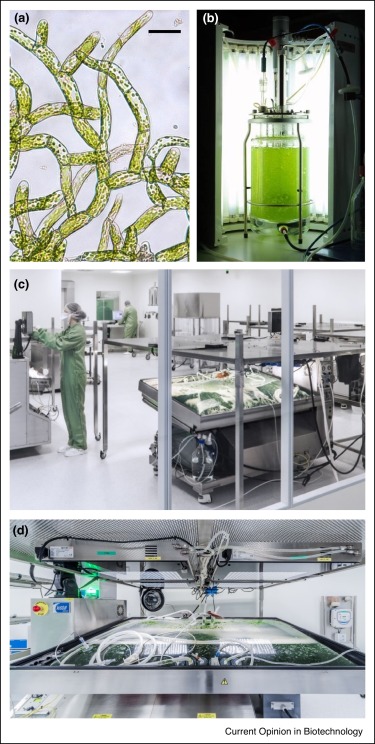 Plant biotechnology often refers to crops and, more recently, algae, but biotechnology also applies to mosses. As Decker and Reski summarize, mosses have some of the same desirable features as algae, including a largely haploid lifecycle that facilitates genetic studies (including homologous recombination and gene editing), and culturability in photobioreactors. To date, moss biotech has produced a cosmetic ingredient, and several high-value pharmaceutical recombinant glycoproteins are being developed, as are protein-engineered biomaterials. Another project has been to produce fragrant moss as room freshners (available now in the US). Outdoors, moss-based biosensors have been developed for environmental monitoring. Finally, biotechnology is being applied to the problem of Sphagnum overharvesting. Because Sphagnum peat moss is an excellent addition to potting soil, it has been overharvested, leading to ecosystem degradation and loss of carbon storage. Fast growth in photobioreactors can accelerate the production of starting material for replacing it in what is described as ‘Sphagnum farming’. (Summary by Mary Williams) Curr. Opin. Biotechnol. 10.1016/j.copbio.2019.09.021
Plant biotechnology often refers to crops and, more recently, algae, but biotechnology also applies to mosses. As Decker and Reski summarize, mosses have some of the same desirable features as algae, including a largely haploid lifecycle that facilitates genetic studies (including homologous recombination and gene editing), and culturability in photobioreactors. To date, moss biotech has produced a cosmetic ingredient, and several high-value pharmaceutical recombinant glycoproteins are being developed, as are protein-engineered biomaterials. Another project has been to produce fragrant moss as room freshners (available now in the US). Outdoors, moss-based biosensors have been developed for environmental monitoring. Finally, biotechnology is being applied to the problem of Sphagnum overharvesting. Because Sphagnum peat moss is an excellent addition to potting soil, it has been overharvested, leading to ecosystem degradation and loss of carbon storage. Fast growth in photobioreactors can accelerate the production of starting material for replacing it in what is described as ‘Sphagnum farming’. (Summary by Mary Williams) Curr. Opin. Biotechnol. 10.1016/j.copbio.2019.09.021
Machine learning enables high-throughput phenotyping for analyses of the genetic architecture of bulliform cell patterning in maize
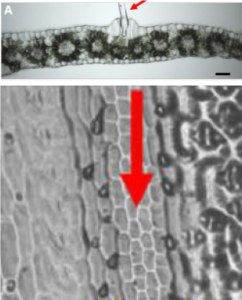 Bulliform cells lie in rows along the upper (adaxial) surface of the maize leaf, and through changes in volume contribute to leaf-rolling, which is a response to water deficit. Several mutants have been identified that affect bulliform cell formation and function, but as yet their occurance in natural populations have not been investigated. One limitation is that microscopic methods are required to identify them, making such analysis labor intensive. Using leaf imprints and machine learning, Qiao et al. developed a high-throughput method to identify bulliform patterning. They used these data as a basis for a GWAS, which when combined with transcriptomic data identified a few candidates for genes involved in bulliform cell patterning. (Summary by Mary Williams) G3 10.1534/g3.119.400757
Bulliform cells lie in rows along the upper (adaxial) surface of the maize leaf, and through changes in volume contribute to leaf-rolling, which is a response to water deficit. Several mutants have been identified that affect bulliform cell formation and function, but as yet their occurance in natural populations have not been investigated. One limitation is that microscopic methods are required to identify them, making such analysis labor intensive. Using leaf imprints and machine learning, Qiao et al. developed a high-throughput method to identify bulliform patterning. They used these data as a basis for a GWAS, which when combined with transcriptomic data identified a few candidates for genes involved in bulliform cell patterning. (Summary by Mary Williams) G3 10.1534/g3.119.400757
Synthetic biogenesis of chromoplasts from leaf chloroplasts
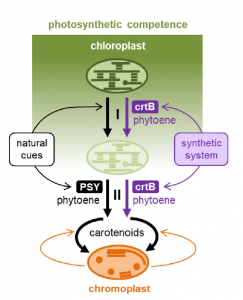 Chromoplasts are a type of plastid, usually found in fruits and flowers, that can accumulate large amounts of carotenoids including beta-carotene (pro-vitamin A). It has been proposed that increasing chromoplast formation could be a way to enhance human consumption of vitamin A. In a new report, Llorente et al. show that they have promoted chloroplast-to-chromoplast transformation in leaves by synthetically overproducing the first committed step of the carotenoid pathway, phytoene synthase. This study shows a good example of how synthetic biology could benefit agriculture and reveals new insights into chromoplast formation. It also demonstrates that loss of photosynthetic competence and overaccumulation of carotenoid are prerequests, not just consequences, of chromoplast differentiation. (Summarized by Nanxun Qin) bioRxiv 10.1101/819177
Chromoplasts are a type of plastid, usually found in fruits and flowers, that can accumulate large amounts of carotenoids including beta-carotene (pro-vitamin A). It has been proposed that increasing chromoplast formation could be a way to enhance human consumption of vitamin A. In a new report, Llorente et al. show that they have promoted chloroplast-to-chromoplast transformation in leaves by synthetically overproducing the first committed step of the carotenoid pathway, phytoene synthase. This study shows a good example of how synthetic biology could benefit agriculture and reveals new insights into chromoplast formation. It also demonstrates that loss of photosynthetic competence and overaccumulation of carotenoid are prerequests, not just consequences, of chromoplast differentiation. (Summarized by Nanxun Qin) bioRxiv 10.1101/819177
Mesostigma viride genome and transcriptome provide insights into the origin and evolution of Streptophyta
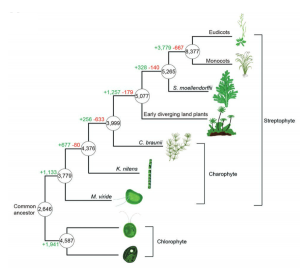 Multicellularity has arisen independently many times across the eukaryotic tree of life (e.g., in plants, animals and fungi). Comparisons of green algae with land plants is likely to identify the genetic toolkit of multicellularity as well as the colonization of terrestrial habitats. Here, Liang et al. present the genome and transcriptome of the single-celled charophyte alga Mesostigma viride, a streptophyte. Streptophytes (single-celled charophytes, multicellular charophytes, and plants) diverged from chlorophyte green algae about a billion years ago. Comparative analyses revealed that M. viride has greater genetic similarity to embryophytes than to unicellular chlorophyte algae. Genes shared between the single-celled M. viride and multicellular plants are associated with multicellular processes such as cell to cell communication and cell division. The research presented provides new insights into the origins of multicellularity and terrestrial flora (Summary by Alex Bowles) Advanced Science 10.1002/advs.201901850
Multicellularity has arisen independently many times across the eukaryotic tree of life (e.g., in plants, animals and fungi). Comparisons of green algae with land plants is likely to identify the genetic toolkit of multicellularity as well as the colonization of terrestrial habitats. Here, Liang et al. present the genome and transcriptome of the single-celled charophyte alga Mesostigma viride, a streptophyte. Streptophytes (single-celled charophytes, multicellular charophytes, and plants) diverged from chlorophyte green algae about a billion years ago. Comparative analyses revealed that M. viride has greater genetic similarity to embryophytes than to unicellular chlorophyte algae. Genes shared between the single-celled M. viride and multicellular plants are associated with multicellular processes such as cell to cell communication and cell division. The research presented provides new insights into the origins of multicellularity and terrestrial flora (Summary by Alex Bowles) Advanced Science 10.1002/advs.201901850
A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels ($)
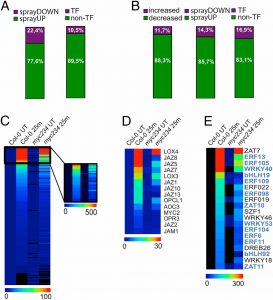 Plants perceive and respond to touch and other physical contact including wounding. Van Moerkercke et al. explored the network of rapid gene responses to a gentle water spray, simulating rainfall. Many of these mecho-induced genes are also responsive to jasmonate, the hormone involved in response to herbivory. A high proportion encode transcription factors (TFs), including a core MYC2/MYC3/MYC4 regulon that is both responsive to jasmonate and also promotes its production in a positive feedback cycle. With tongue in cheek, co-author Harvey Millar reports, “they panic… it seems plants have a fear of rain.” The authors also observe, “this JA- and MYC2/MYC3/MYC4-dependent TF network does not seem to modulate other classic touch marker genes, such as TCH3 and TCH4, meaning that additional touch-induced signaling pathways await discovery.” (Summary by Mary Williams) Proc. Natl. Acad. Sci USA 10.1073/pnas.1911758116
Plants perceive and respond to touch and other physical contact including wounding. Van Moerkercke et al. explored the network of rapid gene responses to a gentle water spray, simulating rainfall. Many of these mecho-induced genes are also responsive to jasmonate, the hormone involved in response to herbivory. A high proportion encode transcription factors (TFs), including a core MYC2/MYC3/MYC4 regulon that is both responsive to jasmonate and also promotes its production in a positive feedback cycle. With tongue in cheek, co-author Harvey Millar reports, “they panic… it seems plants have a fear of rain.” The authors also observe, “this JA- and MYC2/MYC3/MYC4-dependent TF network does not seem to modulate other classic touch marker genes, such as TCH3 and TCH4, meaning that additional touch-induced signaling pathways await discovery.” (Summary by Mary Williams) Proc. Natl. Acad. Sci USA 10.1073/pnas.1911758116
Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome ($)
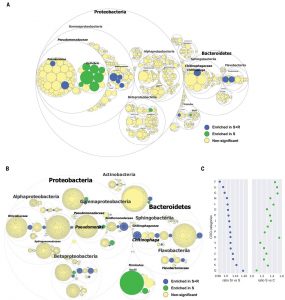 Disease-suppressive soils have long been known, although it hasn’t always been clear how they function. Previous studies have suggested that soil microbes are responsible for disease suppression, because the suppressive property can be transferred to other soils and is lost when soil is sterilized. Carrión et al. grew sugar beet seedlings in suppressive or conducive soils, with or without the fungal pathogen Rhizoctonia solani, and observed a notable reduction in disease in the suppressive soils as expected. They then looked at the microbial composition of root endophytes from plants grown in these conditions and found a few bacterial genera associated with disease suppression, as well as an increased expression of bacterial genes with presumed defensive metabolite and anti-fungal chitinase activities. Based on these results, they introduced a cocktail of candidate species into conducive soils, which led to them becoming suppressive. (Summary by Mary Williams) Science 10.1126/science.aaw9285
Disease-suppressive soils have long been known, although it hasn’t always been clear how they function. Previous studies have suggested that soil microbes are responsible for disease suppression, because the suppressive property can be transferred to other soils and is lost when soil is sterilized. Carrión et al. grew sugar beet seedlings in suppressive or conducive soils, with or without the fungal pathogen Rhizoctonia solani, and observed a notable reduction in disease in the suppressive soils as expected. They then looked at the microbial composition of root endophytes from plants grown in these conditions and found a few bacterial genera associated with disease suppression, as well as an increased expression of bacterial genes with presumed defensive metabolite and anti-fungal chitinase activities. Based on these results, they introduced a cocktail of candidate species into conducive soils, which led to them becoming suppressive. (Summary by Mary Williams) Science 10.1126/science.aaw9285
A mutualistic interaction between Streptomyces bacteria, strawberry plants and pollinating bees
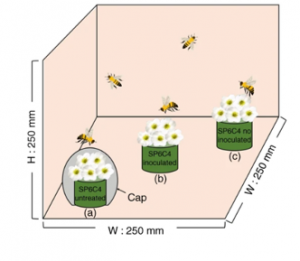 Some species of Streptomyces bacteria produce antimicrobial compounds that have been shown to enhance plant resistance to pathogens. Kim et al. show that his protection can extend to a pollinator. The Streptomyces defends the plant against Botrytis cinerea and protects the bees against insect pathogens (entomopathogens). Furthermore, the beneficial microbe can enter the endosphere and move into the leaves, flowers and pollen, where it can be ingested by the bees and carried on their bodies to other plants (in this case, cultivated strawberries). (Summary by Mary Williams) Nature Comms. 10.1038/s41467-019-12785-3
Some species of Streptomyces bacteria produce antimicrobial compounds that have been shown to enhance plant resistance to pathogens. Kim et al. show that his protection can extend to a pollinator. The Streptomyces defends the plant against Botrytis cinerea and protects the bees against insect pathogens (entomopathogens). Furthermore, the beneficial microbe can enter the endosphere and move into the leaves, flowers and pollen, where it can be ingested by the bees and carried on their bodies to other plants (in this case, cultivated strawberries). (Summary by Mary Williams) Nature Comms. 10.1038/s41467-019-12785-3
Protocol for rapid clearing and staining of fixed Arabidopsis ovules for improved imaging by confocal laser scanning microscopy
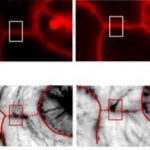
 This is a very interesting paper that provide a new, fast and easy protocol, with specific step-by-step instructions, for a trustworthy imaging with cellular resolution of fixed Arabidopsis ovules at different developmental stages. The authors combine two previously outlined techniques: clearing of fixed ovules in ClearSee solution, and marking the cell outline using the cell wall stain SCRI Renaissance 2200. A key point for the method is the staining with TO-PRO-3 that stains nuclei in fixed ovules, including the nuclei of the embryo sac. Moreover, TO-PRO-3 works well with SR2200, allowing the combination of cell wall and nuclear staining. The best results could be achieved with ClearSee, SR2200 and TO-PRO-3, followed by dissection and mounting the samples in Vectashield. Using the new protocol is possible to generate digital 3D models of ovules of various stages, providing the foundation for a future quantitative analysis of ovule morphogenesis. (Summarized by Francesca Resentini) Plant Methods 10.1186/s13007-019-0505-x
This is a very interesting paper that provide a new, fast and easy protocol, with specific step-by-step instructions, for a trustworthy imaging with cellular resolution of fixed Arabidopsis ovules at different developmental stages. The authors combine two previously outlined techniques: clearing of fixed ovules in ClearSee solution, and marking the cell outline using the cell wall stain SCRI Renaissance 2200. A key point for the method is the staining with TO-PRO-3 that stains nuclei in fixed ovules, including the nuclei of the embryo sac. Moreover, TO-PRO-3 works well with SR2200, allowing the combination of cell wall and nuclear staining. The best results could be achieved with ClearSee, SR2200 and TO-PRO-3, followed by dissection and mounting the samples in Vectashield. Using the new protocol is possible to generate digital 3D models of ovules of various stages, providing the foundation for a future quantitative analysis of ovule morphogenesis. (Summarized by Francesca Resentini) Plant Methods 10.1186/s13007-019-0505-x
Gene-editing in somatic cells can be achieved by spray-on of carbon dots
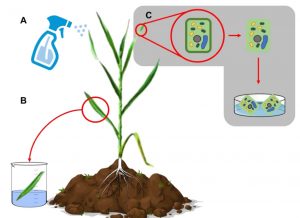 It’s known to all that plant biotechnology can enhance food security, but there are also inevitable disadvantages of these new approaches. At present, introducing DNA into a plant genome is slow and expensive. Recently, Doyle et al. developed a new method which claims to be simple, inexpensive, fast, and versatile in plant species. They mixed carbon nanodots with PEG so that plasmid DNA electrostatically interacts with them. Plants take up these nano-particles delivered via foliar spray. The authors have used this spay-on method to transiently express GFP in the nucleus. Expression of the CRISPR-Cas9 system in somatic cells led to “spray-on gene editing”. The authors demonstrate successful application of this method in wheat, which will be useful for plant developmental and agricultural research. (Summarized by Nanxun Qin) bioRxiv 10.1101/805036
It’s known to all that plant biotechnology can enhance food security, but there are also inevitable disadvantages of these new approaches. At present, introducing DNA into a plant genome is slow and expensive. Recently, Doyle et al. developed a new method which claims to be simple, inexpensive, fast, and versatile in plant species. They mixed carbon nanodots with PEG so that plasmid DNA electrostatically interacts with them. Plants take up these nano-particles delivered via foliar spray. The authors have used this spay-on method to transiently express GFP in the nucleus. Expression of the CRISPR-Cas9 system in somatic cells led to “spray-on gene editing”. The authors demonstrate successful application of this method in wheat, which will be useful for plant developmental and agricultural research. (Summarized by Nanxun Qin) bioRxiv 10.1101/805036
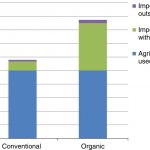
The greenhouse gas impacts of converting food production in England and Wales to organic methods
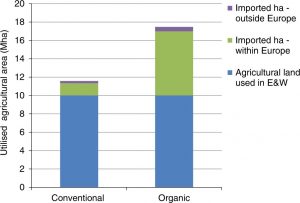 Many discussions about organic food production are dominated by dogma rather than pragmatism. Nevertheless, here’s a useful report that examines the impact of organic farming methods in England and Wales on greenhouse gas (GHG) emissions. Are the well-documented lower yields offset by differences in how the food is produced? Smith et al. suggest not: that when calculated per unit of food, organic results in greater GHG emissions, mainly because the yield penalty must be made up elsewhere. The authors look at all factors affecting net GHG flux, including soil carbon sequestration, production of nitrogen fertilizer, transport, and flame weeding of organic carrots (note: they look at meat and dairy foods as well as plant-based foods). Interestingly, because yields are relatively high in England and Wales as compared to places where the shortfall would be made up, there would need to be a five-fold increase in the amount of overseas land cultivated for food imports. The authors also point out that shifting agricultural production elsewhere simply means moving its inherent impacts, such as decreased biodiversity, elsewhere. Nature Comms. (Summary by Mary Williams) 10.1038/s41467-019-12622-7
Many discussions about organic food production are dominated by dogma rather than pragmatism. Nevertheless, here’s a useful report that examines the impact of organic farming methods in England and Wales on greenhouse gas (GHG) emissions. Are the well-documented lower yields offset by differences in how the food is produced? Smith et al. suggest not: that when calculated per unit of food, organic results in greater GHG emissions, mainly because the yield penalty must be made up elsewhere. The authors look at all factors affecting net GHG flux, including soil carbon sequestration, production of nitrogen fertilizer, transport, and flame weeding of organic carrots (note: they look at meat and dairy foods as well as plant-based foods). Interestingly, because yields are relatively high in England and Wales as compared to places where the shortfall would be made up, there would need to be a five-fold increase in the amount of overseas land cultivated for food imports. The authors also point out that shifting agricultural production elsewhere simply means moving its inherent impacts, such as decreased biodiversity, elsewhere. Nature Comms. (Summary by Mary Williams) 10.1038/s41467-019-12622-7