Plant Science Research Weekly: November 22
Review: The role of peptides cleaved from protein precursors in eliciting plant stress reactions
 Although the first signaling peptide identified in plants, systemin, is involved in stress responses, developmentally important peptide signals have largely occupied the limelight. This Tansley Review by Chen et al. summarizes recent insights into peptides with a role in stress responses: wounding, pathogen infection, nutrient imbalance, drought and high salinity. Many were first identified computationally, and subsequently shown to be biologically active experimentally. Interestingly, only a small subset have been shown to be induced by stress. The review also describes what is known about receptors and signaling events downstream of the peptides. (Summary by Mary Williams) New Phytol. 10.1111/nph.16241
Although the first signaling peptide identified in plants, systemin, is involved in stress responses, developmentally important peptide signals have largely occupied the limelight. This Tansley Review by Chen et al. summarizes recent insights into peptides with a role in stress responses: wounding, pathogen infection, nutrient imbalance, drought and high salinity. Many were first identified computationally, and subsequently shown to be biologically active experimentally. Interestingly, only a small subset have been shown to be induced by stress. The review also describes what is known about receptors and signaling events downstream of the peptides. (Summary by Mary Williams) New Phytol. 10.1111/nph.16241
Review: Ready-to-eat salad crops: A plant pathogen’s heaven
 For those of you celebrating Thanksgiving next weekend, here’s an article full of fun facts to share over the salad. Gullino et al. describe the history of salad (mentioned by Virgil and Pliny) and the rapid growth in the prepared salad industry. They describe the challenge of growing and getting these fragile leaves to market. But they focus on the many pathogens that plague growers; after all, consumers want perfect leaves with no pesticide residues. In addition to many plant pathogens (including Fusarium, Pythium, Sclerotinia, Alternaria and others), human pathogens that infect lettuces are mentioned (you’re sure to be a hit at the dinner table!). Even if you choose not to share your insights with your companions, this is a very interesting and readable article that will make you appreciate the challenges inherent in producing bagged salad, and give thanks to the growers who overcome them. (Summary by Mary Williams) Plant Disease 10.1094/PDIS-03-19-0472-FE
For those of you celebrating Thanksgiving next weekend, here’s an article full of fun facts to share over the salad. Gullino et al. describe the history of salad (mentioned by Virgil and Pliny) and the rapid growth in the prepared salad industry. They describe the challenge of growing and getting these fragile leaves to market. But they focus on the many pathogens that plague growers; after all, consumers want perfect leaves with no pesticide residues. In addition to many plant pathogens (including Fusarium, Pythium, Sclerotinia, Alternaria and others), human pathogens that infect lettuces are mentioned (you’re sure to be a hit at the dinner table!). Even if you choose not to share your insights with your companions, this is a very interesting and readable article that will make you appreciate the challenges inherent in producing bagged salad, and give thanks to the growers who overcome them. (Summary by Mary Williams) Plant Disease 10.1094/PDIS-03-19-0472-FE
Perspective: Revolutionizing agriculture with synthetic biology
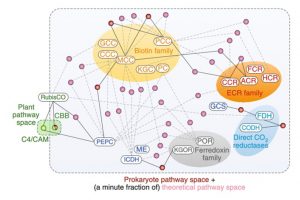 In a new Perspective by Wurtzel et al., the authors lay out SynBio’s tremendous potential to transform agriculture. Consider how we might leverage the “vast design space that plants have not occupied.” As an example, plants employ two pathways to fix carbon, and prokaryotes another six, but scientists have identified another 28 potential pathways (so far) that could be introduced into plants or other organisms, to improve efficiency, resiliency or secondary product formation. Like engineering, synthetic biology invites us to imagine what we’d like to do or have, and then figure out how to make it happen (using engineering’s design-test-build-learn cycle). The authors describe SynBio’s progress in carbon fixation, nitrogen fixation, stress response, and high-value chemical synthesis (e.g., opioids, taxol), and discuss changes needed in “education and training, scientific mindsets, physical infrastructure and societal attitudes”. This latter part has some excellent suggestions for how all plant scientists can contribute towards enabling plant SynBio to flourish and improve our future. (Summary by Mary Williams) Nature Plants 10.1038/s41477-019-0539-0
In a new Perspective by Wurtzel et al., the authors lay out SynBio’s tremendous potential to transform agriculture. Consider how we might leverage the “vast design space that plants have not occupied.” As an example, plants employ two pathways to fix carbon, and prokaryotes another six, but scientists have identified another 28 potential pathways (so far) that could be introduced into plants or other organisms, to improve efficiency, resiliency or secondary product formation. Like engineering, synthetic biology invites us to imagine what we’d like to do or have, and then figure out how to make it happen (using engineering’s design-test-build-learn cycle). The authors describe SynBio’s progress in carbon fixation, nitrogen fixation, stress response, and high-value chemical synthesis (e.g., opioids, taxol), and discuss changes needed in “education and training, scientific mindsets, physical infrastructure and societal attitudes”. This latter part has some excellent suggestions for how all plant scientists can contribute towards enabling plant SynBio to flourish and improve our future. (Summary by Mary Williams) Nature Plants 10.1038/s41477-019-0539-0
Next-Gen sequence databases: RNA and genomic informatics resources for plants
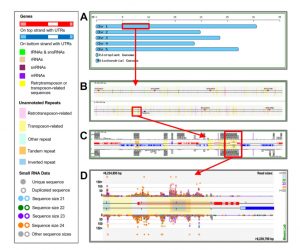 Drawing on more than 15 years of improvements, Nakano et al. have released public websites and resources for data access, display, and analysis of plant small RNAs, from Arabidopsis to wheat and with many crops and model species represented. The tools can analyze, integrate and display the abundance of small RNAs, RNA-Seq and ChIP-Seq data, as well as a new viewer and a query page for the analysis of phased small RNAs. This manuscript introduces these new, improved resources, which are accessible at https://mpss.danforthcenter.org. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.00957
Drawing on more than 15 years of improvements, Nakano et al. have released public websites and resources for data access, display, and analysis of plant small RNAs, from Arabidopsis to wheat and with many crops and model species represented. The tools can analyze, integrate and display the abundance of small RNAs, RNA-Seq and ChIP-Seq data, as well as a new viewer and a query page for the analysis of phased small RNAs. This manuscript introduces these new, improved resources, which are accessible at https://mpss.danforthcenter.org. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.00957
RNA G-quadruplex structures exist and function in vivo
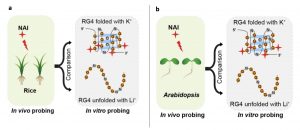 RNA G quadruplex (RG4) structures that include two or more layers of G-quartets can form in guanine-rich RNA sequences. These structures are known exist in vitro and are proposed to form in vivo as well. However, there is no direct evidence from plants showing the presence of these structures in vivo. Here, Yang et al. screened for potential RG4-forming sites and assessed them for their potential to fold as RG4 structures. In silico analysis suggested more than 60,000 potential RG4-forming sites (based on the presence of the GxLnGxLnGxLnGx sequence). RG4 structures cause reverse transcriptional stalling (RTS), and when this functional criterion (rG4-seq) was used fewer than 3000 potential RG4 sites were identified, mostly in coding sequences. The authors determined that the undetected sites are more likely to fold into stable secondary structures rather than RG4 structures. Sites identified in vitro were confirmed in vivo using chemical profiling (SHALiPE-Seq); 2-methylnicotinic acid imidazolide (NAI) preferentially modifies the last Gs of G-tracts when RG4 is folded. They used a genetic approach to confirm the potential role of the RG4 structure in post-transcriptional gene regulation. The existence of RG4 was also confirmed in rice, suggesting the general presence of in vivo RG4s in the plant kingdom. (Summary by Mugdha Sabale) bioRxiv 10.1101/839621
RNA G quadruplex (RG4) structures that include two or more layers of G-quartets can form in guanine-rich RNA sequences. These structures are known exist in vitro and are proposed to form in vivo as well. However, there is no direct evidence from plants showing the presence of these structures in vivo. Here, Yang et al. screened for potential RG4-forming sites and assessed them for their potential to fold as RG4 structures. In silico analysis suggested more than 60,000 potential RG4-forming sites (based on the presence of the GxLnGxLnGxLnGx sequence). RG4 structures cause reverse transcriptional stalling (RTS), and when this functional criterion (rG4-seq) was used fewer than 3000 potential RG4 sites were identified, mostly in coding sequences. The authors determined that the undetected sites are more likely to fold into stable secondary structures rather than RG4 structures. Sites identified in vitro were confirmed in vivo using chemical profiling (SHALiPE-Seq); 2-methylnicotinic acid imidazolide (NAI) preferentially modifies the last Gs of G-tracts when RG4 is folded. They used a genetic approach to confirm the potential role of the RG4 structure in post-transcriptional gene regulation. The existence of RG4 was also confirmed in rice, suggesting the general presence of in vivo RG4s in the plant kingdom. (Summary by Mugdha Sabale) bioRxiv 10.1101/839621
The genome of the Charophyte alga Penium margaritaceum bears footprints of the evolutionary origins of land plants
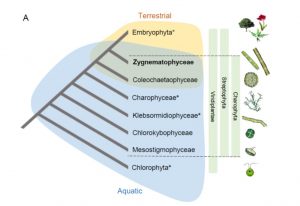 The Zygnematophyceae are the green algae that are most closely related to land plants. Some species in this clade are considered subaerial, meaning that they can live under air (as opposed to under water). The green films you see on tree trunks and walls are often Zygnematophycaea. Several new insights into the terrestrialization of land plants have emerged from genome sequences and functional analyses of this group of near-plant relatives. In a new preprint, Jiao, Sørensen, and Sun share insights from Penium margaritaceum, a single-celled member of the Zygnematophyceae. In this genome, the authors found abundant transposable elements that might have contributed to genetic innovations, and evidence for the emergence and amplification of gene families associated with terrestrialization. Among these are the machinery for remodeling the cell walls into those suitable for life on land. (Summary by Mary Williams) bioRxiv 10.1101/835561
The Zygnematophyceae are the green algae that are most closely related to land plants. Some species in this clade are considered subaerial, meaning that they can live under air (as opposed to under water). The green films you see on tree trunks and walls are often Zygnematophycaea. Several new insights into the terrestrialization of land plants have emerged from genome sequences and functional analyses of this group of near-plant relatives. In a new preprint, Jiao, Sørensen, and Sun share insights from Penium margaritaceum, a single-celled member of the Zygnematophyceae. In this genome, the authors found abundant transposable elements that might have contributed to genetic innovations, and evidence for the emergence and amplification of gene families associated with terrestrialization. Among these are the machinery for remodeling the cell walls into those suitable for life on land. (Summary by Mary Williams) bioRxiv 10.1101/835561
Genomes of subaerial Zygnematophyceae provide insights into land plant evolution ($)
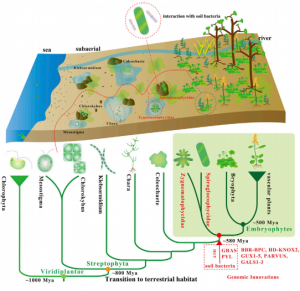 The availability of charophyte algae genomic information is helping to understand how the plant transition to a terrestrial environment occurred at the molecular level. Here, Cheng et al. are releasing two genomes from the Zygnematophyceae clade (Spirogloea muscicola and Mesotaenium endlicherianum), among the first genomes from this clade, the closest to most land plants (embryophytes). In contrast to other charophytes, these species can live outside the water environment. As it was deduced before from transcriptomes, key components of the genetic toolkit associated with land life have originated in the common ancestor to this clade and embryophytes. Interestingly, this paper sheds light on the origin of the ABA receptors (PYL) and GRAS transcription factors, suggesting they came from a horizontal gene transfer (HGT) event from soil bacteria. They propose that the common ancestor to embryophytes and Zygnematophyceae lived in a terrestrial environment. This study raises questions about the hypothesis that most genetic innovations evolved in the aquatic environment, and opens a scenario where plants gained genes from HGT while being on the land but before the radiation of embryophytes. (Summary by Facundo Romani) Cell 10.1016/j.cell.2019.10.019
The availability of charophyte algae genomic information is helping to understand how the plant transition to a terrestrial environment occurred at the molecular level. Here, Cheng et al. are releasing two genomes from the Zygnematophyceae clade (Spirogloea muscicola and Mesotaenium endlicherianum), among the first genomes from this clade, the closest to most land plants (embryophytes). In contrast to other charophytes, these species can live outside the water environment. As it was deduced before from transcriptomes, key components of the genetic toolkit associated with land life have originated in the common ancestor to this clade and embryophytes. Interestingly, this paper sheds light on the origin of the ABA receptors (PYL) and GRAS transcription factors, suggesting they came from a horizontal gene transfer (HGT) event from soil bacteria. They propose that the common ancestor to embryophytes and Zygnematophyceae lived in a terrestrial environment. This study raises questions about the hypothesis that most genetic innovations evolved in the aquatic environment, and opens a scenario where plants gained genes from HGT while being on the land but before the radiation of embryophytes. (Summary by Facundo Romani) Cell 10.1016/j.cell.2019.10.019
A ligand-independent origin of abscisic acid perception ($)
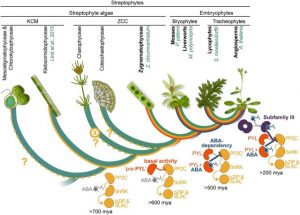 The origins of ABA signaling have long been puzzling. Green algae make ABA but don’t respond to it, and ABA receptors have appeared to be missing from their genomes. Reccently, Sun et al. identified a gene encoding an ortholog of PYL family of ABA receptor proteins in the transcriptome of an alga from the Zygnematophyceae clade (closest relatives to land plants), and here they asses it functionally. They like other researchers note the similarity of the Zygnematophyceae algae to land plants and their apparent predisposition to life on land. Their key finding is that in these algae PYL-like proteins interact with PP2C-like proteins, but in an ABA-independent manner. They propose, “that co-option of ABA to control a preexisting PP2C-SnRK2-dependent desiccation-tolerance pathway enabled transition from an all-or-nothing survival strategy to a hormone-modulated, competitive strategy by enabling continued growth of anatomically diversifying vascular plants in dehydrative conditions.” (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA
The origins of ABA signaling have long been puzzling. Green algae make ABA but don’t respond to it, and ABA receptors have appeared to be missing from their genomes. Reccently, Sun et al. identified a gene encoding an ortholog of PYL family of ABA receptor proteins in the transcriptome of an alga from the Zygnematophyceae clade (closest relatives to land plants), and here they asses it functionally. They like other researchers note the similarity of the Zygnematophyceae algae to land plants and their apparent predisposition to life on land. Their key finding is that in these algae PYL-like proteins interact with PP2C-like proteins, but in an ABA-independent manner. They propose, “that co-option of ABA to control a preexisting PP2C-SnRK2-dependent desiccation-tolerance pathway enabled transition from an all-or-nothing survival strategy to a hormone-modulated, competitive strategy by enabling continued growth of anatomically diversifying vascular plants in dehydrative conditions.” (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA
Emergence of the Ug99 lineage of the wheat stem rust pathogen through somatic hybridization
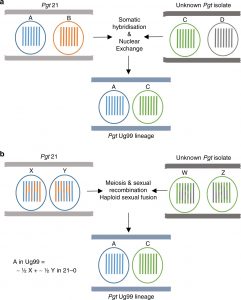 The Ug99 strain of wheat stem rust pathogen Puccinia graminis f. sp. tritici (Pgt) emerged in Uganda in 1999 and presents a significant threat to global wheat production. Genetic analysis indicates it is distinct from other Pgt races. Li et al. set out to understand its origins. Puccinia graminis are dikaryotic fungi meaning they carry two distinct haploid nuclei. In the absence of their alternate host they usually reproduce asexualy, limiting opportunities to increase their genetic diversity. Ug99 carries one haploid nucleus from a known isolate, Pgt21, and another from an unknown donor, and there has been no recombination between these genomes. Thus, Ug99 is derived from the nonsexual process of somatic hybridization. “These findings indicate that nuclear exchange between dikaryotes can generate genetic diversity and facilitate the emergence of new lineages in asexual fungal populations.” (Summary by Mary Williams) Nature Comms. 10.1038/s41467-019-12927-7
The Ug99 strain of wheat stem rust pathogen Puccinia graminis f. sp. tritici (Pgt) emerged in Uganda in 1999 and presents a significant threat to global wheat production. Genetic analysis indicates it is distinct from other Pgt races. Li et al. set out to understand its origins. Puccinia graminis are dikaryotic fungi meaning they carry two distinct haploid nuclei. In the absence of their alternate host they usually reproduce asexualy, limiting opportunities to increase their genetic diversity. Ug99 carries one haploid nucleus from a known isolate, Pgt21, and another from an unknown donor, and there has been no recombination between these genomes. Thus, Ug99 is derived from the nonsexual process of somatic hybridization. “These findings indicate that nuclear exchange between dikaryotes can generate genetic diversity and facilitate the emergence of new lineages in asexual fungal populations.” (Summary by Mary Williams) Nature Comms. 10.1038/s41467-019-12927-7
Pollination of Cretaceous flowers
 Like something from Jurassic Park, a tiny insect embedded in amber has provided new insights into life millions of years ago. But in this case, the 99 million year old insect shows us that, as Darwin surmised, insects really were important contributors to angiosperm pollination from their origins. In fact, this pollen-carrying insect (a tumbling flower beetle) pushes back the known date for insect pollination of angiosperms by 50 million years (previously, only insect pollination of gymnosperms was known from this time). By using X-ray microcomputed tomography (micro-CT) and confocal microscopy, Bao et al. show that the amber-preserved beetle has pollen-carrying hairs, and a body shape and mouth parts that are consistent with pollination. Their key finding is that it is carrying tricolpate (three-grooved) pollen typical of eudicots, truly a validating insects’ contribution in early angiosperm success. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1916186116
Like something from Jurassic Park, a tiny insect embedded in amber has provided new insights into life millions of years ago. But in this case, the 99 million year old insect shows us that, as Darwin surmised, insects really were important contributors to angiosperm pollination from their origins. In fact, this pollen-carrying insect (a tumbling flower beetle) pushes back the known date for insect pollination of angiosperms by 50 million years (previously, only insect pollination of gymnosperms was known from this time). By using X-ray microcomputed tomography (micro-CT) and confocal microscopy, Bao et al. show that the amber-preserved beetle has pollen-carrying hairs, and a body shape and mouth parts that are consistent with pollination. Their key finding is that it is carrying tricolpate (three-grooved) pollen typical of eudicots, truly a validating insects’ contribution in early angiosperm success. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1916186116



