Plant Science Research Weekly: May 27th, 2022
Systematic characterization of gene function in the photosynthetic alga Chlamydomonas reinhardtii
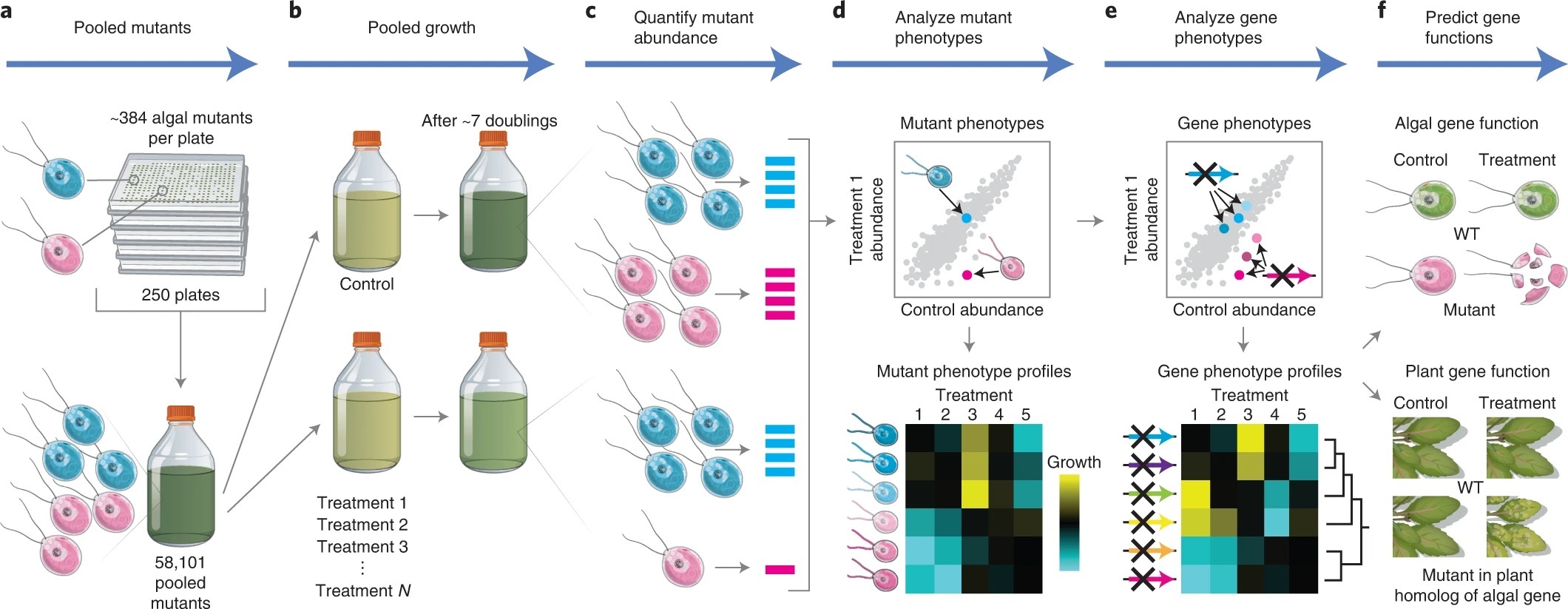 The green alga Chlamydomonas reinhardtii is a useful model system to study photosynthetic organisms, as this single-celled species allows for more high-throughput methods than in multicellular plants, and many conserved pathways of interest can be identified. Here, Fauser et al. demonstrate the value of these methods in a phenotypic screen of a previously generated barcoded insertion mutant library of Chlamydomonas. They used pooled cultures from a total of 58101 mutants covering 14695 genes, under 121 environmental and chemical stress conditions, using difference in growth as the assessed phenotype. They were able to make specific associations between genotypes and phenotypes and demonstrated the robustness of this method by confirming known genetic pathways, by clustering genes based on similar phenotypes. This also allowed them to characterise potential new players in various genetic pathways, such as the kinase MARS1 as playing a part in the activation of the chloroplast unfolded protein response upon DNA damage. They also identified three new genes in the defence pathway against actin inhibition by soil microorganisms (LAT5, LAT6, and LAT7) and showed conservation of function of these genes in Arabidopsis through mutant studies. Other characterised pathways include photosynthesis, CO2 concentrating mechanism, and cilia function, providing a useful resource for guiding gene characterisation in plants. (Summary by Jiawen Chen @jiaaawen ) Nature Genetics 10.1038/s41588-022-01052-9
The green alga Chlamydomonas reinhardtii is a useful model system to study photosynthetic organisms, as this single-celled species allows for more high-throughput methods than in multicellular plants, and many conserved pathways of interest can be identified. Here, Fauser et al. demonstrate the value of these methods in a phenotypic screen of a previously generated barcoded insertion mutant library of Chlamydomonas. They used pooled cultures from a total of 58101 mutants covering 14695 genes, under 121 environmental and chemical stress conditions, using difference in growth as the assessed phenotype. They were able to make specific associations between genotypes and phenotypes and demonstrated the robustness of this method by confirming known genetic pathways, by clustering genes based on similar phenotypes. This also allowed them to characterise potential new players in various genetic pathways, such as the kinase MARS1 as playing a part in the activation of the chloroplast unfolded protein response upon DNA damage. They also identified three new genes in the defence pathway against actin inhibition by soil microorganisms (LAT5, LAT6, and LAT7) and showed conservation of function of these genes in Arabidopsis through mutant studies. Other characterised pathways include photosynthesis, CO2 concentrating mechanism, and cilia function, providing a useful resource for guiding gene characterisation in plants. (Summary by Jiawen Chen @jiaaawen ) Nature Genetics 10.1038/s41588-022-01052-9
CLAVATA3/ESR-related 2 peptides regulate root sucrose level and root/shoot ratio in Arabidopsis
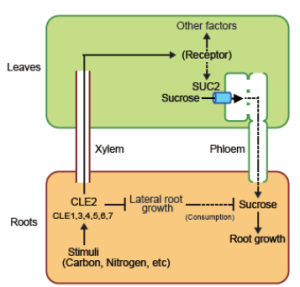 Roots are dependent on shoot-produced sucrose for energy associated with growth and nutrient uptake. The CLAVATA3/ESR2-related peptides (CLEs) regulate several aspects of plant growth. Okamoto et al. present evidence on the involvement of CLEs in conveying the root sucrose demand to the shoot. Carbon starvation upregulates CLE2 and CLE3 and increases the translocation of CLE2 from root to shoot. Transient ectopic expression of CLE2 and overexpression of CLE2 in vascular tissues increases the root sucrose content. The septuple cle mutant, Atcle1-7, is deficient in root sucrose status. Under diurnally fluctuating as well as normal light conditions, shoot/root mass was significantly higher in Atcle1-7 septuple mutants in comparison to wild type. The expression of the sucrose/H+ transporter SUC2 (involved in sucrose phloem loading) was lower in Atcle1-7 septuple mutants. Application of synthetic CLE2 peptide could rescue root sucrose content of Atcle1-7 mutants and partially rescue SUC2 expression. Reciprocal grafting of Atcle1-7 with wild type roots and shoots revealed that root genotype of CLE could regulate shoot SUC2 gene expression. Thus, CLEs are root-derived signals that pass information to shoots and determine root sucrose content. (Summary by Lekshmy Sathee @lekshmysnair) Plant Physiol. 10.1093/plphys/kiac227
Roots are dependent on shoot-produced sucrose for energy associated with growth and nutrient uptake. The CLAVATA3/ESR2-related peptides (CLEs) regulate several aspects of plant growth. Okamoto et al. present evidence on the involvement of CLEs in conveying the root sucrose demand to the shoot. Carbon starvation upregulates CLE2 and CLE3 and increases the translocation of CLE2 from root to shoot. Transient ectopic expression of CLE2 and overexpression of CLE2 in vascular tissues increases the root sucrose content. The septuple cle mutant, Atcle1-7, is deficient in root sucrose status. Under diurnally fluctuating as well as normal light conditions, shoot/root mass was significantly higher in Atcle1-7 septuple mutants in comparison to wild type. The expression of the sucrose/H+ transporter SUC2 (involved in sucrose phloem loading) was lower in Atcle1-7 septuple mutants. Application of synthetic CLE2 peptide could rescue root sucrose content of Atcle1-7 mutants and partially rescue SUC2 expression. Reciprocal grafting of Atcle1-7 with wild type roots and shoots revealed that root genotype of CLE could regulate shoot SUC2 gene expression. Thus, CLEs are root-derived signals that pass information to shoots and determine root sucrose content. (Summary by Lekshmy Sathee @lekshmysnair) Plant Physiol. 10.1093/plphys/kiac227
Spermine inhibits PAMP ROS and Ca2+ burst and reshapes the transcriptional landscape of PAMP-triggered immunity in Arabidopsis
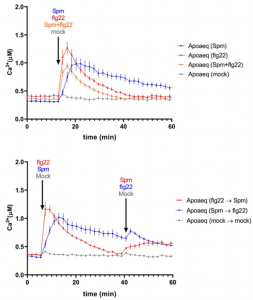 Polyamines are endogenous cationic amines that have been shown to regulate a plethora of biological processes in plants, including the hypersensitive response. However, the importance of polyamines in other aspects of plant immunity has not yet been fully explored. In this new preprint, Zhang and colleagues examine the role of two polyamines, spermine and putrescine, in various aspects of defence in Arabidopsis thaliana. First, the group showed that treatment with exogenous spermine induced a small but sustained ROS wave, but inhibited the characteristic ROS burst associated with exposure to the bacterial peptide flagellin. This effect was independent of salicylic acid signalling pathways but was dependent on the NADPH oxidase RBOHD. The researchers took a pharmacological approach to dissect this mechanism, concluding that spermine inhibits ROS production via direct inhibition of RBOHD or its ROS scavenging ability, rather than via its interactions with the flagellin receptor. Interestingly spermine treatment also induced cytosolic calcium influx, but reduced the calcium increase induced by flagellin, suggesting that spermine may block RBOHD activation via its upstream effects on calcium signalling. Finally, the group conducted gene expression analysis on plants treated with combinations of the two polyamines and flagellin, showing that both polyamines can alter defence associated transcriptional programs. (Summary by Rory Burke @rorby95) bioRxiv 10.1101/2022.05.05.490826
Polyamines are endogenous cationic amines that have been shown to regulate a plethora of biological processes in plants, including the hypersensitive response. However, the importance of polyamines in other aspects of plant immunity has not yet been fully explored. In this new preprint, Zhang and colleagues examine the role of two polyamines, spermine and putrescine, in various aspects of defence in Arabidopsis thaliana. First, the group showed that treatment with exogenous spermine induced a small but sustained ROS wave, but inhibited the characteristic ROS burst associated with exposure to the bacterial peptide flagellin. This effect was independent of salicylic acid signalling pathways but was dependent on the NADPH oxidase RBOHD. The researchers took a pharmacological approach to dissect this mechanism, concluding that spermine inhibits ROS production via direct inhibition of RBOHD or its ROS scavenging ability, rather than via its interactions with the flagellin receptor. Interestingly spermine treatment also induced cytosolic calcium influx, but reduced the calcium increase induced by flagellin, suggesting that spermine may block RBOHD activation via its upstream effects on calcium signalling. Finally, the group conducted gene expression analysis on plants treated with combinations of the two polyamines and flagellin, showing that both polyamines can alter defence associated transcriptional programs. (Summary by Rory Burke @rorby95) bioRxiv 10.1101/2022.05.05.490826
Robust transcriptional indicators of immune cell death revealed by spatio-temporal
transcriptome analyses
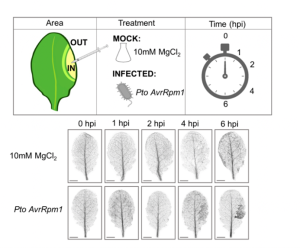 Many plants trigger a form of cell death, known as the hypersensitive response (HR), immediately upon pathogen recognition. To minimize damage to the plant, this cell death must be highly localized to the site of attack, while more distal cells survive and activate other immune responses. However, the transcriptomic programs underlying these disparate cell fates have yet to be uncovered. Here, Salguero-Linares et al. utilized an Arabidopsis thaliana–Pseudomonas syringae system and RNA-sequencing technology to isolate genes specifically implicated in hypersensitive cell death. By comparing the transcriptomes of infected areas with that of tissue outside of the inoculation zone across 5 different time points, the group were able to identify 13 marker genes for HR. The researchers then generated transgenic plants expressing green fluorescence protein under the promoters of each of the 13 genes. One line, corresponding to an uncharacterised AAA-ATPase, showed a specific fluorescence signal only in the pathogen inoculated areas destined to undergo HR, indicating this is a marker for cells destined to die. Interestingly, these plants also produced the same signals following inoculation with other bacteria which induce HR via recognition by different types of plant immune receptors, but not when inoculated with non-pathogenic bacteria, indicating that this reporter line may serve as a reliable tool for further studies of this form of plant immunity. (Summary by Rory Burke @rorby95) Mol. Plant 10.1016/j.molp.2022.04.010
Many plants trigger a form of cell death, known as the hypersensitive response (HR), immediately upon pathogen recognition. To minimize damage to the plant, this cell death must be highly localized to the site of attack, while more distal cells survive and activate other immune responses. However, the transcriptomic programs underlying these disparate cell fates have yet to be uncovered. Here, Salguero-Linares et al. utilized an Arabidopsis thaliana–Pseudomonas syringae system and RNA-sequencing technology to isolate genes specifically implicated in hypersensitive cell death. By comparing the transcriptomes of infected areas with that of tissue outside of the inoculation zone across 5 different time points, the group were able to identify 13 marker genes for HR. The researchers then generated transgenic plants expressing green fluorescence protein under the promoters of each of the 13 genes. One line, corresponding to an uncharacterised AAA-ATPase, showed a specific fluorescence signal only in the pathogen inoculated areas destined to undergo HR, indicating this is a marker for cells destined to die. Interestingly, these plants also produced the same signals following inoculation with other bacteria which induce HR via recognition by different types of plant immune receptors, but not when inoculated with non-pathogenic bacteria, indicating that this reporter line may serve as a reliable tool for further studies of this form of plant immunity. (Summary by Rory Burke @rorby95) Mol. Plant 10.1016/j.molp.2022.04.010
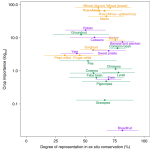
State of ex situ conservation of landrace groups of 25 major crops
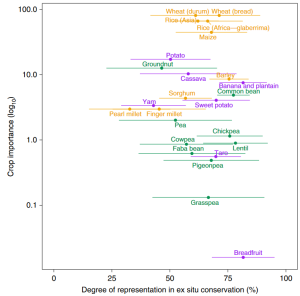 Landraces are local cultivars developed by traditional agricultural methods. Landraces not only represent an essential genetic resource for modern crop breeding but are also key to understand crops’ domestication processes. Thus, it is crucial to collect landraces for genebank conservation since demographic changes and climate emergency threaten in situ populations. Although there have been extensive efforts for landraces to be represented ex situ (landraces are more conserved than their wild relatives), there is a limited information about the distribution of landraces and their degree of representation in genebanks. In this work, Ramirez-Villegas et al., use a conservation gap analysis methodology that predicts the distributions and quantify the current ex situ conservation of more than 70 groups of landraces. At the landrace group level, differences in conservation status were found within many crops depending on morphologic characteristics. To conclude, the modelling processes and the landrace group classification described highlight the potential of associating characteristics of cultivated plant populations with environmental and socioeconomic predictors within the origin and the crop diversity. In any case, the authors also emphasize the necessity of field reconnaissance and of partnering with traditional agrarian communities. (Summary by Eva Maria Gomez Alvarez, @eva_ga96). Nature Plants. 10.1038/s41477-022-01144-8
Landraces are local cultivars developed by traditional agricultural methods. Landraces not only represent an essential genetic resource for modern crop breeding but are also key to understand crops’ domestication processes. Thus, it is crucial to collect landraces for genebank conservation since demographic changes and climate emergency threaten in situ populations. Although there have been extensive efforts for landraces to be represented ex situ (landraces are more conserved than their wild relatives), there is a limited information about the distribution of landraces and their degree of representation in genebanks. In this work, Ramirez-Villegas et al., use a conservation gap analysis methodology that predicts the distributions and quantify the current ex situ conservation of more than 70 groups of landraces. At the landrace group level, differences in conservation status were found within many crops depending on morphologic characteristics. To conclude, the modelling processes and the landrace group classification described highlight the potential of associating characteristics of cultivated plant populations with environmental and socioeconomic predictors within the origin and the crop diversity. In any case, the authors also emphasize the necessity of field reconnaissance and of partnering with traditional agrarian communities. (Summary by Eva Maria Gomez Alvarez, @eva_ga96). Nature Plants. 10.1038/s41477-022-01144-8