Plant Science Research Weekly: March 7
Review: How mycorrhizal associations drive plant population and community biology ($)
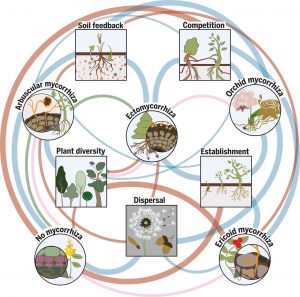 Great strides have been made in discovering the molecular players that allow plants and mycorrhizal fungi to establish their symbiosis. Here, Tedersoo et al. look beyond the single plant and address how these associations affect plant communities. Notably, they review the functions of the four evolutionarily distinct types of mycorrhizal associations separately and identify commonalities and distinctions: arbuscular mycorrhiza (AM), ectomycorrhiza (EcM), ericoid mycorrhiza (ErM), and orchid mycorrhiza (OM). In addition to the well-described contribution to nutrient uptake, mycorrhizal associations can facilitate communication between plants through the “common mycelial network”, condition soil, and confer protection against pathogens. Competition and cooperation outcomes can differ between kin and non-kin, as well as within and between species. The authors categorize the impacts on community ecology as deriving from effects on plant dispersal, establishment, and coexistence. (Summary by Mary Williams) Science 10.1126/science.aba1223
Great strides have been made in discovering the molecular players that allow plants and mycorrhizal fungi to establish their symbiosis. Here, Tedersoo et al. look beyond the single plant and address how these associations affect plant communities. Notably, they review the functions of the four evolutionarily distinct types of mycorrhizal associations separately and identify commonalities and distinctions: arbuscular mycorrhiza (AM), ectomycorrhiza (EcM), ericoid mycorrhiza (ErM), and orchid mycorrhiza (OM). In addition to the well-described contribution to nutrient uptake, mycorrhizal associations can facilitate communication between plants through the “common mycelial network”, condition soil, and confer protection against pathogens. Competition and cooperation outcomes can differ between kin and non-kin, as well as within and between species. The authors categorize the impacts on community ecology as deriving from effects on plant dispersal, establishment, and coexistence. (Summary by Mary Williams) Science 10.1126/science.aba1223
Review: Mechanisms regulating PIF transcription factor activity at the protein level ($)
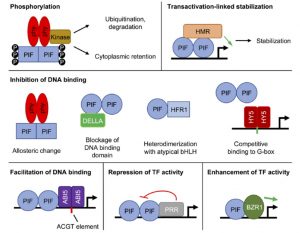 Absorption of red and far-red light by phytochromes dramatically affects plant development, and many of these effects are mediated by the PIF (PHYTOCHROME INTERACTING FACTOR) family of transcription factors. Favero reviews the many ways that PIF activity is regulated beyond through their interactions with phytochrome. These include post-translational modifications leading to protein turnover, sequestration into photobodies, and interactions with other proteins (dimerization, heterodimerization, and competition) that affect PIF stability, binding to DNA and activity. Reading this review highlights the near ubiquity of PIFs in plant processes; guest appearances are made by key components of many other signaling pathways spanning the circadian clock to growth-regulatory hormones. Many researchers will have some familiarity with one or more PIF effects, but it’s particullarly useful to have them all described in one place for a birds-eye view of this important family of transcriptional regulators. (Summary by Mary Williams) Physiol. Plant. 10.1111/ppl.13075
Absorption of red and far-red light by phytochromes dramatically affects plant development, and many of these effects are mediated by the PIF (PHYTOCHROME INTERACTING FACTOR) family of transcription factors. Favero reviews the many ways that PIF activity is regulated beyond through their interactions with phytochrome. These include post-translational modifications leading to protein turnover, sequestration into photobodies, and interactions with other proteins (dimerization, heterodimerization, and competition) that affect PIF stability, binding to DNA and activity. Reading this review highlights the near ubiquity of PIFs in plant processes; guest appearances are made by key components of many other signaling pathways spanning the circadian clock to growth-regulatory hormones. Many researchers will have some familiarity with one or more PIF effects, but it’s particullarly useful to have them all described in one place for a birds-eye view of this important family of transcriptional regulators. (Summary by Mary Williams) Physiol. Plant. 10.1111/ppl.13075
High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize
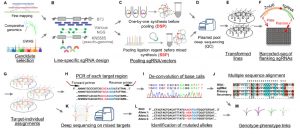 Maize has provided some fascinating mutants and developmental insights, but its genomic complexity has made it more difficult (for example as compared to rice) to identify agronomically important genes. Liu et al. describe a new high-throughput method to integrate forward- and reverse-genetics to identify genes in maize and other crops with complex genomes. They selected 1244 potentially interesting candidate genes that had been identified from either classical mapping or comparative genomics. They made six CRISPR libraries of sgRNAs, which they transformed into maize embryos using Agrobacterium, and sequenced the transformants using primers from the conserved plasmid backbone to identify induced sequence variation. Sixty percent of the changes were small deletions (1-65 bp), with the remainder being small insertions or base substitutions. They found only a small number of off-target mutations, but a high rate of mosaicism. The data produced from this work will help others design efficient mutagenesis approaches and anticipate the outcomes, and, the “library of edited genes provides an unprecedented resource for further detailed functional genomics.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.0093
Maize has provided some fascinating mutants and developmental insights, but its genomic complexity has made it more difficult (for example as compared to rice) to identify agronomically important genes. Liu et al. describe a new high-throughput method to integrate forward- and reverse-genetics to identify genes in maize and other crops with complex genomes. They selected 1244 potentially interesting candidate genes that had been identified from either classical mapping or comparative genomics. They made six CRISPR libraries of sgRNAs, which they transformed into maize embryos using Agrobacterium, and sequenced the transformants using primers from the conserved plasmid backbone to identify induced sequence variation. Sixty percent of the changes were small deletions (1-65 bp), with the remainder being small insertions or base substitutions. They found only a small number of off-target mutations, but a high rate of mosaicism. The data produced from this work will help others design efficient mutagenesis approaches and anticipate the outcomes, and, the “library of edited genes provides an unprecedented resource for further detailed functional genomics.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.0093
Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms
 The trait of herbicide tolerance allows farmers to use chemical means to eliminate weed competitors. Acetolactate synthase (ALS) is an enzyme targeted by more than 50 different herbicides. In order to generate novel herbicide tolerance traits, Kuang et al. used a base-editing artificial evolution approach, by targeting nucleotide-modifying enzymes (cytosine deaminase and adenosine deaminase) to the gene using the CRISPR/Cas9 system. 63 sgRNAs spanning the rice ALS1 open reading frame were introduced into base-editing constructs, which were introduced into rice through ballistics or Agrobacterium-mediated transformation. Tissues were regenerated without herbicide selection to assay for overall gene editing efficiency, and in the presence of the herbicide to identify evolved tolerance. One new variant, which conferred particularly strong tolerance without a yield penalty, was easily introduced into another elite cultivar though gene editing. (Summary by Mary Williams) Mol. Plant 10.1016/j.molp.2020.01.010
The trait of herbicide tolerance allows farmers to use chemical means to eliminate weed competitors. Acetolactate synthase (ALS) is an enzyme targeted by more than 50 different herbicides. In order to generate novel herbicide tolerance traits, Kuang et al. used a base-editing artificial evolution approach, by targeting nucleotide-modifying enzymes (cytosine deaminase and adenosine deaminase) to the gene using the CRISPR/Cas9 system. 63 sgRNAs spanning the rice ALS1 open reading frame were introduced into base-editing constructs, which were introduced into rice through ballistics or Agrobacterium-mediated transformation. Tissues were regenerated without herbicide selection to assay for overall gene editing efficiency, and in the presence of the herbicide to identify evolved tolerance. One new variant, which conferred particularly strong tolerance without a yield penalty, was easily introduced into another elite cultivar though gene editing. (Summary by Mary Williams) Mol. Plant 10.1016/j.molp.2020.01.010
Structural basis of unidirectional energy transfer in Porphyridium purpureum phycobilisome
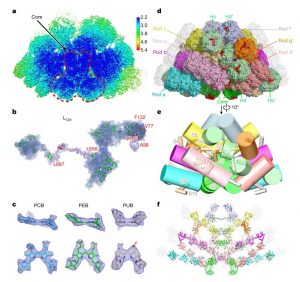 Cyanobacteria and red algae employ phycobilisomes (PBSs) as light-harvesting systems to adapt to fluctuating environments. PBSs are composed of phycobiliproteins, linker proteins and chromophores. Ma et al. used cryo-EM to determine the 2.82 Å structure of the very large (14.7-megadalton, 706 protein subunit) PBS from the unicellular reg alga Porphyridium purpureum, which they compared to that of a previously characterized PBS from the multicellular red alga Griffithsia pacifica. The newly characterized PBS complex contained 528 phycoerythrin, 72 phycocyanin, 46 allophycocyanin, 60 linker proteins and 1598 chromophores. The structure provides insights into how the energy states of the chromophores can be affected through interacting with the aromatic amino acids of the linker proteins, therefore ensuring the efficient unidirectional transfer of energy within phycobilisome. The study shows how light is harvested and transferred in the PBS of cyanobacteria and red algae, and will benefit further study of artificial photosynthesis. (Summary by Nanxun Qin) Nature 10.1038/s41586-020-2020-7
Cyanobacteria and red algae employ phycobilisomes (PBSs) as light-harvesting systems to adapt to fluctuating environments. PBSs are composed of phycobiliproteins, linker proteins and chromophores. Ma et al. used cryo-EM to determine the 2.82 Å structure of the very large (14.7-megadalton, 706 protein subunit) PBS from the unicellular reg alga Porphyridium purpureum, which they compared to that of a previously characterized PBS from the multicellular red alga Griffithsia pacifica. The newly characterized PBS complex contained 528 phycoerythrin, 72 phycocyanin, 46 allophycocyanin, 60 linker proteins and 1598 chromophores. The structure provides insights into how the energy states of the chromophores can be affected through interacting with the aromatic amino acids of the linker proteins, therefore ensuring the efficient unidirectional transfer of energy within phycobilisome. The study shows how light is harvested and transferred in the PBS of cyanobacteria and red algae, and will benefit further study of artificial photosynthesis. (Summary by Nanxun Qin) Nature 10.1038/s41586-020-2020-7
Chimeric activators and repressors define HY5 activity and reveal a light-regulated feedback mechanism
 Plants take cues from the environment and decide when and how to regulate growth. When light is limiting, etiolated growth allows plants to reach the soil surface and gain access to light. After light perception, plants de-etiolate and go through a series of morphological and genetic changes. HY5 is one of the key transcription factors regulating de-etiolation and photomorphogenesis, although its function in transcription has been unclear. Burko et al. analyzed fusion proteins of HY5 with either a constitutive transcriptional activator (VP16) or repressor (SRDX) in Arabidopsis and tomato. Expressing HY5-VP16 and overexpressing HY5 rescue the long hypocotyl of hy5 seedlings whereas expressing HY5-SRDX exacerbates many hy5 mutant phenotypes. RNA-seq data show that in genes with high confidence HY5 binding sites (based on ChIP-seq), 79% are upregulated in HY5-VP16 and downregulated in HY5-SRDX, consistent with HY5 mostly working as a transcriptional activator during de-etiolation, and further identifying its direct targets. The study also demonstrates, “a molecular feedback loop via the COP1/SPA E3-ubiquitin ligase complex suggesting a mechanism which maintains low HY5 in the dark, primed for rapid accumulation to reprogram growth upon light exposure.” This approach also provide a promising strategy for future transcription factor research. (Summary by Yun-Ting Kao) Plant Cell 10.1105/tpc.19.00772
Plants take cues from the environment and decide when and how to regulate growth. When light is limiting, etiolated growth allows plants to reach the soil surface and gain access to light. After light perception, plants de-etiolate and go through a series of morphological and genetic changes. HY5 is one of the key transcription factors regulating de-etiolation and photomorphogenesis, although its function in transcription has been unclear. Burko et al. analyzed fusion proteins of HY5 with either a constitutive transcriptional activator (VP16) or repressor (SRDX) in Arabidopsis and tomato. Expressing HY5-VP16 and overexpressing HY5 rescue the long hypocotyl of hy5 seedlings whereas expressing HY5-SRDX exacerbates many hy5 mutant phenotypes. RNA-seq data show that in genes with high confidence HY5 binding sites (based on ChIP-seq), 79% are upregulated in HY5-VP16 and downregulated in HY5-SRDX, consistent with HY5 mostly working as a transcriptional activator during de-etiolation, and further identifying its direct targets. The study also demonstrates, “a molecular feedback loop via the COP1/SPA E3-ubiquitin ligase complex suggesting a mechanism which maintains low HY5 in the dark, primed for rapid accumulation to reprogram growth upon light exposure.” This approach also provide a promising strategy for future transcription factor research. (Summary by Yun-Ting Kao) Plant Cell 10.1105/tpc.19.00772
Two MYB proteins in a self-organizing activator-inhibitor system produce spotted pigmentation patterns
 The questions of how patterns are formed is one of the oldest in biology, and even considered by the famous mathematician Alan Turing, who proposed that reaction-diffusion (RD) models underly de novo pattern formation. Briefly, a reaction that takes place in one place sends a signal that leads to a different effect. A new paper identifies two genes involved in a RD model for the origins of pollinator-attracting nectar guides in the monkeyflower Mimulus spp. NECTAR GUIDE ANTHOCYANIN (NEGAN) is a MYB transcription factor in monkeyflower necessary for anthocyanin deposition in nectar-guide spots. In this new work, the authors speculated that the spots could be generated through a Turing-type RD pattern generator. Through a genetic screen for plants in which the entire nectar-guide region was red (rather than spotted), they identified a mutant in a MYB-encoding gene that they named RED TONGUE, with a function of inhbiting anthocyanin production. They also found natural variants with the same phenotype caused by the same gene in a related species, Mimulus guttatus. NEGAN activates expression of RTO (consistent with an RD mechanism). Furthermore, RTO is a negative regulator of NEGAN function that moves between cells. Computer simulations confirm the potential of this gene pair to produce localized spots of anthocyanin. Finally, the authors also found allelic variation in wild populations, and showed that the corresponding phenotypic variation is capable of affecting pollinator visits. (Summary by Mary Williams) Curr. Biol. 10.1016/j.cub.2019.12.067
The questions of how patterns are formed is one of the oldest in biology, and even considered by the famous mathematician Alan Turing, who proposed that reaction-diffusion (RD) models underly de novo pattern formation. Briefly, a reaction that takes place in one place sends a signal that leads to a different effect. A new paper identifies two genes involved in a RD model for the origins of pollinator-attracting nectar guides in the monkeyflower Mimulus spp. NECTAR GUIDE ANTHOCYANIN (NEGAN) is a MYB transcription factor in monkeyflower necessary for anthocyanin deposition in nectar-guide spots. In this new work, the authors speculated that the spots could be generated through a Turing-type RD pattern generator. Through a genetic screen for plants in which the entire nectar-guide region was red (rather than spotted), they identified a mutant in a MYB-encoding gene that they named RED TONGUE, with a function of inhbiting anthocyanin production. They also found natural variants with the same phenotype caused by the same gene in a related species, Mimulus guttatus. NEGAN activates expression of RTO (consistent with an RD mechanism). Furthermore, RTO is a negative regulator of NEGAN function that moves between cells. Computer simulations confirm the potential of this gene pair to produce localized spots of anthocyanin. Finally, the authors also found allelic variation in wild populations, and showed that the corresponding phenotypic variation is capable of affecting pollinator visits. (Summary by Mary Williams) Curr. Biol. 10.1016/j.cub.2019.12.067
Hybrid autoimmunity and a plant resistosome complex ($)
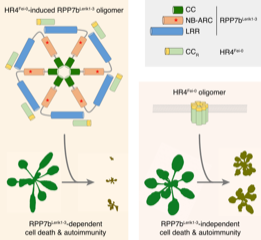 Hybrid necrosis occurs when the progeny of a cross between two different plants show widespread cell death. It can be caused by autoactivation of nucleotide-binding and leucine rich repeat domain (NLR) proteins, intracellular immune receptors that play a central role in plant resistance to diverse pathogens. Li et al. showed that hybrid necrosis in the cross between A. thaliana accessions Lerik1-3 and Fei-0 is caused when the NLR RPP7 from Lerik1-3 (RPP7b) and the non-NLR HR4 from Fei-0 (HR4Fei-0) are co-expressed in a single plant. They showed that HR4Fei-0 interacts with RPP7b and promotes RPP7b to form a higher-order complex in plants; this oligomerization and subsequent cell death activation require the P-loop and leucine-rich repeat (LRR) domain of RPP7b. Interestingly, HR4Fei-0 forms self-oligomers and triggers autoimmunity in plants in an RPP7b-independent manner. Also, HR4Fei-0, when expressed in bacteria, can kill bacterial cells in the absence of other plant NLRs. A mutant of HR4 compromised in self-oligomerization and RPP7b-independent cell death was also compromised in RPP7b-dependent cell death. Based on the results, the authors proposed a model in which HR4 oligomers, potentially mimicking HR4 targeted by pathogen effectors, act as ligands that activate RPP7b through promoting this NLR to form oligomers. This study demonstrates that hybrid necrosis can be used to mechanistically interrogate key components of the plant immune system. (Summary by Tatsuya Nobori) Cell Host Microbe 10.1016/j.chom.2020.01.012
Hybrid necrosis occurs when the progeny of a cross between two different plants show widespread cell death. It can be caused by autoactivation of nucleotide-binding and leucine rich repeat domain (NLR) proteins, intracellular immune receptors that play a central role in plant resistance to diverse pathogens. Li et al. showed that hybrid necrosis in the cross between A. thaliana accessions Lerik1-3 and Fei-0 is caused when the NLR RPP7 from Lerik1-3 (RPP7b) and the non-NLR HR4 from Fei-0 (HR4Fei-0) are co-expressed in a single plant. They showed that HR4Fei-0 interacts with RPP7b and promotes RPP7b to form a higher-order complex in plants; this oligomerization and subsequent cell death activation require the P-loop and leucine-rich repeat (LRR) domain of RPP7b. Interestingly, HR4Fei-0 forms self-oligomers and triggers autoimmunity in plants in an RPP7b-independent manner. Also, HR4Fei-0, when expressed in bacteria, can kill bacterial cells in the absence of other plant NLRs. A mutant of HR4 compromised in self-oligomerization and RPP7b-independent cell death was also compromised in RPP7b-dependent cell death. Based on the results, the authors proposed a model in which HR4 oligomers, potentially mimicking HR4 targeted by pathogen effectors, act as ligands that activate RPP7b through promoting this NLR to form oligomers. This study demonstrates that hybrid necrosis can be used to mechanistically interrogate key components of the plant immune system. (Summary by Tatsuya Nobori) Cell Host Microbe 10.1016/j.chom.2020.01.012
Mildew Locus O facilitates colonization by arbuscular mycorrhizal fungi in angiosperms
 In plants, disease resistance genes typically act in a dominant way – the presence of a resistance allele, even a single copy, is enough to confer resistance. The barley gene Mildew Resistance Locus O (MLO1) is different, as it acts in a recessive way; loss-of-function mlo1 plants are resistant to the pathogenic powdery mildew fungus. Therefore, the presence of MLO1 confers disease susceptibility. Jacott et al. set out to ask why plant genomes carry a gene that makes them more susceptible to disease. Interestingly, they found that mlo1 mutants are also less able to form associations with mycorrhizal fungi, demonstrating a positive benefit for the MLO1 gene, which the authors suggest has been exploited by the pathogen. (Summary by Mary Williams) New Phytol. 10.1111/nph.16465
In plants, disease resistance genes typically act in a dominant way – the presence of a resistance allele, even a single copy, is enough to confer resistance. The barley gene Mildew Resistance Locus O (MLO1) is different, as it acts in a recessive way; loss-of-function mlo1 plants are resistant to the pathogenic powdery mildew fungus. Therefore, the presence of MLO1 confers disease susceptibility. Jacott et al. set out to ask why plant genomes carry a gene that makes them more susceptible to disease. Interestingly, they found that mlo1 mutants are also less able to form associations with mycorrhizal fungi, demonstrating a positive benefit for the MLO1 gene, which the authors suggest has been exploited by the pathogen. (Summary by Mary Williams) New Phytol. 10.1111/nph.16465
Some mycoheterotrophic orchids depend on carbon from deadwood: novel evidence from a radiocarbon approach
 A mycoheterotrophic (“fungal-other-eating”) plant takes carbon nutrients from a fungus, but as fungi are not themselves photosynthetic, the (ectomycorrhizal) fungus must get its carbon from somewhere, usually a plant. Thus the typical flow of carbon goes from autotrophic photosynthesizing plant to fungus to mycoheterotrophic plant. Now, using carbon dating, Suetsugu et al. have shown that some mycoheterotrophic orchids get their carbon from (saprotrophic) fungi that get their carbon from decaying plant tissues, in this case deadwood. Although 13C carbon isotope analysis has been used previously to identify sources of photosynthetic carbon, in this case the study focused on 14C analysis to determine the age of the carbon source. As described by the authors, “The radiocarbon approach is powerful because the Δ14C values of atmospheric CO2 were globally elevated by nuclear weapons testing in the early 1950s, but have steadily declined since the nuclear test ban treaty of 1963.” The authors further discuss the use of 14C-enriched “bomb carbon” to address the nutritional mode of plants. (Summary by Mary Williams) New Phytol. 10.1111/nph.16409
A mycoheterotrophic (“fungal-other-eating”) plant takes carbon nutrients from a fungus, but as fungi are not themselves photosynthetic, the (ectomycorrhizal) fungus must get its carbon from somewhere, usually a plant. Thus the typical flow of carbon goes from autotrophic photosynthesizing plant to fungus to mycoheterotrophic plant. Now, using carbon dating, Suetsugu et al. have shown that some mycoheterotrophic orchids get their carbon from (saprotrophic) fungi that get their carbon from decaying plant tissues, in this case deadwood. Although 13C carbon isotope analysis has been used previously to identify sources of photosynthetic carbon, in this case the study focused on 14C analysis to determine the age of the carbon source. As described by the authors, “The radiocarbon approach is powerful because the Δ14C values of atmospheric CO2 were globally elevated by nuclear weapons testing in the early 1950s, but have steadily declined since the nuclear test ban treaty of 1963.” The authors further discuss the use of 14C-enriched “bomb carbon” to address the nutritional mode of plants. (Summary by Mary Williams) New Phytol. 10.1111/nph.16409
Community diversity outweighs effect of warming on plant colonization ($)
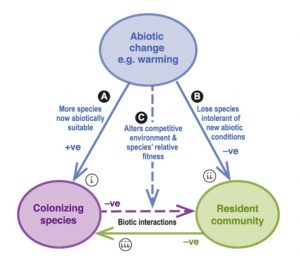 Several studies have addressed growing concerns regarding increases in exotic plant colonization of communities in response to a warming climate. However, both abiotic (e.g., warming climate) and biotic (e.g., changes in community diversity) shifts can be simultaneous results of warming and both can influence colonization of a community. The question that a recent experiment by Catford et al. aimed to address was whether one type of shift was more important than the other for colonizer success. Grassland plots were seeded and then warmed with infrared heaters. Some plots were maintained at the original species richness, or number of diverse species, while others were not. After 20 years, biomass samples were collected and colonizing species (non-seeded) were quantified. Separating abiotic and biotic influences in this way showed that high diversity communities and communities with particular functional (physical) traits could resist colonization by certain types of species despite warming of up to 3°C, suggesting that biotic shifts in response to warming may be more important than abiotic shifts for invaders. (Summary by Sienna Wessel) Global Change Biology 10.1111/gcb.15017
Several studies have addressed growing concerns regarding increases in exotic plant colonization of communities in response to a warming climate. However, both abiotic (e.g., warming climate) and biotic (e.g., changes in community diversity) shifts can be simultaneous results of warming and both can influence colonization of a community. The question that a recent experiment by Catford et al. aimed to address was whether one type of shift was more important than the other for colonizer success. Grassland plots were seeded and then warmed with infrared heaters. Some plots were maintained at the original species richness, or number of diverse species, while others were not. After 20 years, biomass samples were collected and colonizing species (non-seeded) were quantified. Separating abiotic and biotic influences in this way showed that high diversity communities and communities with particular functional (physical) traits could resist colonization by certain types of species despite warming of up to 3°C, suggesting that biotic shifts in response to warming may be more important than abiotic shifts for invaders. (Summary by Sienna Wessel) Global Change Biology 10.1111/gcb.15017