Plant Science Research Weekly: March 5, 2021
Review: Perspectives on improving light distribution and light use efficiency in crop canopies
 Light comes largely from above, and plants have evolved strategies to efficiently capture this light even when competing with other potentially shading plants. However, at a field-level, such competition may (and does) prevent plants from collectively maximizing light use efficiency, hence yield. In this comprehensive review, Slatterly and Ott provide an overview of years of study into how light use efficiency could be optimized in crop plants. These strategies include alterations in plant architecture and leaf size/position, as well as changing photosynthetic pigment properties (e.g., red-shifting), to allow more light to reach into lower canopy layers. (Summary by Mary Williams @PlantTeaching) Plant Physiol 10.1093/plphys/kiaa006. Don’t miss the excellent accompanying profile “Agriculture futurist: Don Ort” written by Assistant Features Editor Meisha Holloway-Phillips. Plant Physiol. 10.1093/plphys/kiaa024
Light comes largely from above, and plants have evolved strategies to efficiently capture this light even when competing with other potentially shading plants. However, at a field-level, such competition may (and does) prevent plants from collectively maximizing light use efficiency, hence yield. In this comprehensive review, Slatterly and Ott provide an overview of years of study into how light use efficiency could be optimized in crop plants. These strategies include alterations in plant architecture and leaf size/position, as well as changing photosynthetic pigment properties (e.g., red-shifting), to allow more light to reach into lower canopy layers. (Summary by Mary Williams @PlantTeaching) Plant Physiol 10.1093/plphys/kiaa006. Don’t miss the excellent accompanying profile “Agriculture futurist: Don Ort” written by Assistant Features Editor Meisha Holloway-Phillips. Plant Physiol. 10.1093/plphys/kiaa024
Genome assembly and population genomic analysis provide insights into the evolution of modern sweet corn
 In maize (Zea mays), loss of function mutations in genes involved in starch biosynthesis characterize sweet corn varieties that have increased sugar content in the kernel. Specifically, lines carrying the shrunken2 (sh2) allele revolutionized the corn industry in Northern America in the last 50 years thanks to improved quality traits and shelf life. Besides the kernel, these cultivars also display several phenotypic alterations compared to field corn, suggesting they originated through divergent selection. In this paper, Hu and coworkers presented a high-quality reference genome of an agronomically important sweet corn inbred line (la435) harboring the sh2-R allele (Ia453-sh2). Results of Transposable Elements (TE) annotation and pairwise genomic comparison with six field corn varieties, including the B73 reference genome, indicated high level of divergence in TE families among accessions, which could be related to genetic variation in protein-coding genes. As an example, the sweet corn la453-sh2 defective allele is associated with structural rearrangements causing a 50 Kb separation between the first and the second half of the gene. This big TE insertion – which is absent in B73 – caused the loss of a conserved domain in the enzyme that controls the first irreversible reaction of starch biosynthesis, leading to higher sugar accumulation in the endosperm. This paper presents novel insights into the phylogenetic relationships among more than 5000 maize accessions and identifies candidate genes likely to control additional sweet corn traits such as leaf architecture (Drooping leaf), short stature (Dwarf1) and early flowering (FT-interacting protein). (Summary and image adaptation by Michela Osnato @michela_osnato). Nature Comms. 10.1038/s41467-021-21380-4
In maize (Zea mays), loss of function mutations in genes involved in starch biosynthesis characterize sweet corn varieties that have increased sugar content in the kernel. Specifically, lines carrying the shrunken2 (sh2) allele revolutionized the corn industry in Northern America in the last 50 years thanks to improved quality traits and shelf life. Besides the kernel, these cultivars also display several phenotypic alterations compared to field corn, suggesting they originated through divergent selection. In this paper, Hu and coworkers presented a high-quality reference genome of an agronomically important sweet corn inbred line (la435) harboring the sh2-R allele (Ia453-sh2). Results of Transposable Elements (TE) annotation and pairwise genomic comparison with six field corn varieties, including the B73 reference genome, indicated high level of divergence in TE families among accessions, which could be related to genetic variation in protein-coding genes. As an example, the sweet corn la453-sh2 defective allele is associated with structural rearrangements causing a 50 Kb separation between the first and the second half of the gene. This big TE insertion – which is absent in B73 – caused the loss of a conserved domain in the enzyme that controls the first irreversible reaction of starch biosynthesis, leading to higher sugar accumulation in the endosperm. This paper presents novel insights into the phylogenetic relationships among more than 5000 maize accessions and identifies candidate genes likely to control additional sweet corn traits such as leaf architecture (Drooping leaf), short stature (Dwarf1) and early flowering (FT-interacting protein). (Summary and image adaptation by Michela Osnato @michela_osnato). Nature Comms. 10.1038/s41467-021-21380-4
Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes
 During domestication of maize, one of the favorable traits was meristem size. Understanding the genetic circuit of maize meristem development and engineering for crop productivity are important from an agricultural perspective. In this article, Lie et al. used CRISPR-Cas9 to edit the promoter regions of ZmCLE7 (ZmCLAVATA3/EMBRYO SURROUNDING REGION-RELATED7), ZmFCP1 (ZmFON2-LIKE CLE PROTEIN1) and coding regions of ZmCLE1E5 to generating weak or null alleles, respectively. They eventually generated 13 alleles: six for ZmCLE7, five for ZmFCP1, and two for ZmCLE1E5. The promoter edited alleles of ZmCLE7 and coding region-edited alleles of ZmCLE1E5 have larger inflorescence meristem (IMs) diameters. Additionally, the authors found that these alleles have increased kernel row numbers and grain yields. This work highlights CRIPSR-Cas9-mediated gene editing as a tool to modify agriculturally important traits such as meristem size in maize. (Summary by Arif Ashraf) Nature Plants 10.1038/s41477-021-00858-5
During domestication of maize, one of the favorable traits was meristem size. Understanding the genetic circuit of maize meristem development and engineering for crop productivity are important from an agricultural perspective. In this article, Lie et al. used CRISPR-Cas9 to edit the promoter regions of ZmCLE7 (ZmCLAVATA3/EMBRYO SURROUNDING REGION-RELATED7), ZmFCP1 (ZmFON2-LIKE CLE PROTEIN1) and coding regions of ZmCLE1E5 to generating weak or null alleles, respectively. They eventually generated 13 alleles: six for ZmCLE7, five for ZmFCP1, and two for ZmCLE1E5. The promoter edited alleles of ZmCLE7 and coding region-edited alleles of ZmCLE1E5 have larger inflorescence meristem (IMs) diameters. Additionally, the authors found that these alleles have increased kernel row numbers and grain yields. This work highlights CRIPSR-Cas9-mediated gene editing as a tool to modify agriculturally important traits such as meristem size in maize. (Summary by Arif Ashraf) Nature Plants 10.1038/s41477-021-00858-5
Molecular mechanism of cytokinin-activated cell division in Arabidopsis
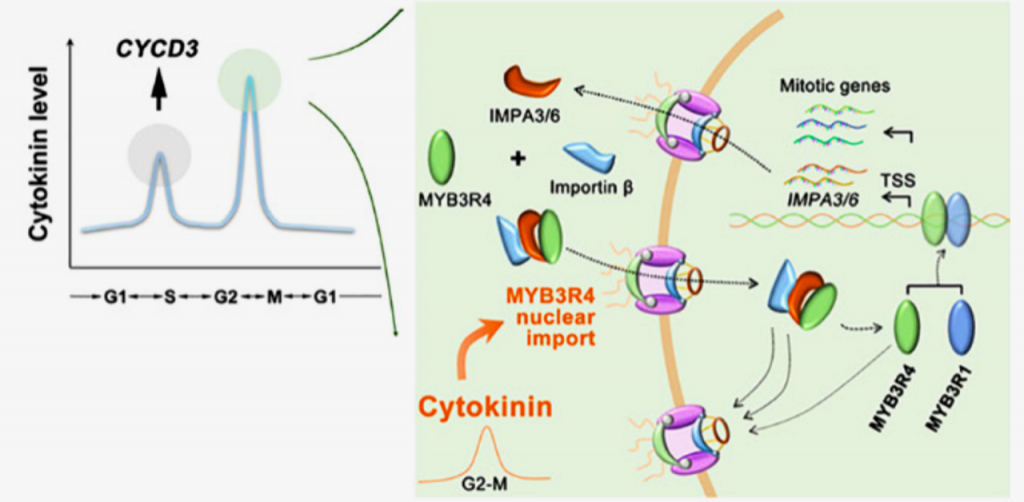 Proper organogenesis depends on the precise control of cell proliferation and differentiation. Cytokinin and auxin have emerged as key regulators of both these processes, where cytokinin promotes proliferation and synergistically with auxin fine-tunes the proliferation and differentiation rate. Years of research and evidence have implicated cytokinin as a central regulator of stem cell proliferation in the Arabidopsis shoot apical meristem (SAM). Although the precise molecular mechanisms underlying proliferation and cytokinin signaling have been well studied, how cytokinin promotes mitotic cell division is obscure. A recent paper by Yang and colleagues identified the MYB family transcription factor MYB3R4 as a key player controlling cytokinin-induced mitotic cell division. The authors propose a mechanism in which high cytokinin levels at the G2/M transition promote the rapid translocation of MYB3R4 from the cytoplasm to the nucleus, where MYB3R4 in cooperation with MYB3R1 activates a mitotic gene expression cascade to drive cell division and cytokinesis. This gene expression cascade is reinforced by a feedforward loop involving two direct targets of MYB3R4/MYB3R1, importins IMPA3 and IMPA6, which facilitates faster and stronger MYB3R4 nuclear trafficking. Furthermore, the authors highlight the importance of MYB3R4 nuclear localization as evidenced by the fact that a constitutively nuclear localized form of MYB3R4 can partially phenocopy cytokinin action. This work coupled with previous research into cytokinin-induced cell division provides a mechanistic understanding of how different levels of cytokinin throughout the cell cycle modulate cell division and thus precise control of stem cell number in the SAM. (Summary by Jesus Leon @jesussaur) Science 10.1126/science.abe2305
Proper organogenesis depends on the precise control of cell proliferation and differentiation. Cytokinin and auxin have emerged as key regulators of both these processes, where cytokinin promotes proliferation and synergistically with auxin fine-tunes the proliferation and differentiation rate. Years of research and evidence have implicated cytokinin as a central regulator of stem cell proliferation in the Arabidopsis shoot apical meristem (SAM). Although the precise molecular mechanisms underlying proliferation and cytokinin signaling have been well studied, how cytokinin promotes mitotic cell division is obscure. A recent paper by Yang and colleagues identified the MYB family transcription factor MYB3R4 as a key player controlling cytokinin-induced mitotic cell division. The authors propose a mechanism in which high cytokinin levels at the G2/M transition promote the rapid translocation of MYB3R4 from the cytoplasm to the nucleus, where MYB3R4 in cooperation with MYB3R1 activates a mitotic gene expression cascade to drive cell division and cytokinesis. This gene expression cascade is reinforced by a feedforward loop involving two direct targets of MYB3R4/MYB3R1, importins IMPA3 and IMPA6, which facilitates faster and stronger MYB3R4 nuclear trafficking. Furthermore, the authors highlight the importance of MYB3R4 nuclear localization as evidenced by the fact that a constitutively nuclear localized form of MYB3R4 can partially phenocopy cytokinin action. This work coupled with previous research into cytokinin-induced cell division provides a mechanistic understanding of how different levels of cytokinin throughout the cell cycle modulate cell division and thus precise control of stem cell number in the SAM. (Summary by Jesus Leon @jesussaur) Science 10.1126/science.abe2305
Mechanism and function of root circumnutation
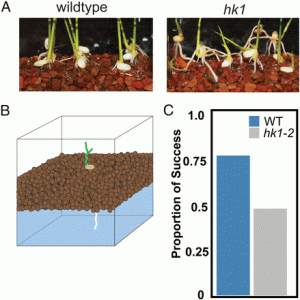 This is an engrossing paper. The authors start off with a simple mutant screen for abnormal root growth in rice seedlings, through which they identified three “long root” mutant alleles of a rice mutant in a gene encoding a histidine kinase, HK1. Careful analysis showed that the mutant roots fail to circumnutate (a growth pattern derived from helical movement at the tip). Part of this paper is a conventional analysis of the molecular underpinning of this circumnutation, revealing roles for ethylene and intercellular auxin movement, but the other part is an intriguing quest to understand the function of circumnutation. For this, the authors developed a robotic root, which allowed them to test how helical tip movements affect growth. These studies support a model in which a non-circumnutating root that hits an obstacle “head on” will get stuck, whereas a circumnutating tip will successfully go around obstacles (clearly shown in this video). This model is further supported from growth assays using the hk1 mutant; wild-type roots can push through gravelly substrates, whereas the non-circumnutating roots cannot, leading to a failure of the seedling to successfully anchor to the substrate (again, watch the video). This would be a great paper to read with students, as both the question and findings are straightforward, novel, and interesting. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2018940118
This is an engrossing paper. The authors start off with a simple mutant screen for abnormal root growth in rice seedlings, through which they identified three “long root” mutant alleles of a rice mutant in a gene encoding a histidine kinase, HK1. Careful analysis showed that the mutant roots fail to circumnutate (a growth pattern derived from helical movement at the tip). Part of this paper is a conventional analysis of the molecular underpinning of this circumnutation, revealing roles for ethylene and intercellular auxin movement, but the other part is an intriguing quest to understand the function of circumnutation. For this, the authors developed a robotic root, which allowed them to test how helical tip movements affect growth. These studies support a model in which a non-circumnutating root that hits an obstacle “head on” will get stuck, whereas a circumnutating tip will successfully go around obstacles (clearly shown in this video). This model is further supported from growth assays using the hk1 mutant; wild-type roots can push through gravelly substrates, whereas the non-circumnutating roots cannot, leading to a failure of the seedling to successfully anchor to the substrate (again, watch the video). This would be a great paper to read with students, as both the question and findings are straightforward, novel, and interesting. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2018940118
Phytoplasma effector physically interacts with host proteasome to promote bacterial growth
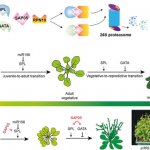
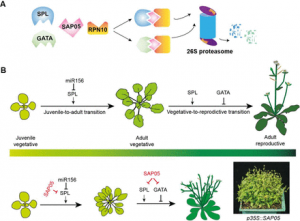 As a way to circumnavigate the plant innate immune system, phytopathogens evolved effector molecules that protect the pathogen from the plant’s defenses. SAP05, an effector from the insect-vectored phytoplasma Candidatus (a bacterial obligate parasite), is responsible for the plant phenotype observed in witches broom disease, characterized by increased vegetative growth and disrupted reproductive growth. In this exciting new research, Huang and colleagues investigate the SAP05 method of action, and demonstrate that SAP05 directly interacts with Arabidopsis ubiquitin receptor, AtRPN10, as well as members of the host transcription factor (TF) families, GATA and SPL, responsible for mediating growth and development. This paper demonstrates a novel destabilization strategy wherein SAP05 bridges GATA or SPL and AtRPN10 in a ubiquitin-independent manner, allowing RPN10 to destabilize the TFs. The simultaneous destabilization of GATA and SPL TF families causes the decoupling of developmental transitions in infected plants, allowing rampant bacterial proliferation in plant tissues. Despite the highly conserved sequence of RPN10 within eukaryotes, SAP05 targets two sequential lowly-conserved residues in AtPRN10. Mutation of AtRPN10 to resemble that of the insect vector prevents SAP05 binding, and effectively confers resistance to the SAP05 effector. In elucidating this novel mechanism, this paper provides viable avenues for therapeutic degradation of target proteins, as well as options for improving plant resistance to phytoplasma pathogens. (Summary by Marlo Hall @marloXplants). bioRxiv 10.1101/2021.02.15.430920
As a way to circumnavigate the plant innate immune system, phytopathogens evolved effector molecules that protect the pathogen from the plant’s defenses. SAP05, an effector from the insect-vectored phytoplasma Candidatus (a bacterial obligate parasite), is responsible for the plant phenotype observed in witches broom disease, characterized by increased vegetative growth and disrupted reproductive growth. In this exciting new research, Huang and colleagues investigate the SAP05 method of action, and demonstrate that SAP05 directly interacts with Arabidopsis ubiquitin receptor, AtRPN10, as well as members of the host transcription factor (TF) families, GATA and SPL, responsible for mediating growth and development. This paper demonstrates a novel destabilization strategy wherein SAP05 bridges GATA or SPL and AtRPN10 in a ubiquitin-independent manner, allowing RPN10 to destabilize the TFs. The simultaneous destabilization of GATA and SPL TF families causes the decoupling of developmental transitions in infected plants, allowing rampant bacterial proliferation in plant tissues. Despite the highly conserved sequence of RPN10 within eukaryotes, SAP05 targets two sequential lowly-conserved residues in AtPRN10. Mutation of AtRPN10 to resemble that of the insect vector prevents SAP05 binding, and effectively confers resistance to the SAP05 effector. In elucidating this novel mechanism, this paper provides viable avenues for therapeutic degradation of target proteins, as well as options for improving plant resistance to phytoplasma pathogens. (Summary by Marlo Hall @marloXplants). bioRxiv 10.1101/2021.02.15.430920