Plant Science Research Weekly: March 20
Update: How plants sense and respond to stressful environments
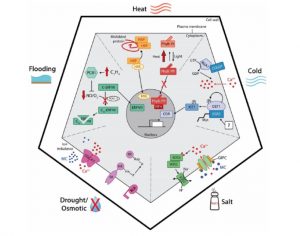 A longstanding question in plant science is how plants “know” that they are under threat. The identification of cell-surface receptors that identify conserved pathogen patterns sheds some light on biotic stress perception, but what about abiotic stresses such as excessive heat or drought? Lamers et al. give an update on what we currently understand about how different environmental factors are sensed (or not) and how these signals transduced into responses. As an example, the light sensor phytochrome B has been recently identified as key heat sensor that converts from active to inactive form at a rate that is sensitive to temperature. By contrast, although several cold-responsive proteins have been identified, none stand out as the key primary sensor at this point. Membrane fluidity, which is affected by temperature, is not considered to be a primary signal for temperature responses. Sensing mechanisms of drought, flooding and excess salinity are also reviewed. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.01464
A longstanding question in plant science is how plants “know” that they are under threat. The identification of cell-surface receptors that identify conserved pathogen patterns sheds some light on biotic stress perception, but what about abiotic stresses such as excessive heat or drought? Lamers et al. give an update on what we currently understand about how different environmental factors are sensed (or not) and how these signals transduced into responses. As an example, the light sensor phytochrome B has been recently identified as key heat sensor that converts from active to inactive form at a rate that is sensitive to temperature. By contrast, although several cold-responsive proteins have been identified, none stand out as the key primary sensor at this point. Membrane fluidity, which is affected by temperature, is not considered to be a primary signal for temperature responses. Sensing mechanisms of drought, flooding and excess salinity are also reviewed. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.01464
Perspective: Multiscale computational models can guide experimentation and targeted measurements for crop improvement
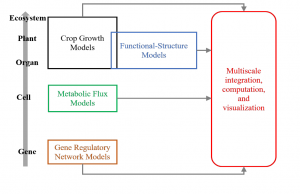 Throughout the plant science community, the use of computational or in silico analyses which precede traditional studies are gaining traction to identify research opportunities. Multiscale computational models are those which assimilate data from all biological system levels from gene to ecosystem. Benes et al. advocate for the use of such multiscale models to advise crop improvement studies. The authors present examples in which single scale mathematical models have previously guided research such as to inform crop management strategies. However, by combining models of different biological scales, we can evaluate crop “ideotypes” and test “what if” scenarios to comprehend how crops will react in untried conditions. To that end, we need to develop methods to bridge independently designed models, which is a major aim of the Crops in silico consortium. This group has developed the yggdrasil framework which facilitates connections between models that operate at distinct biological scales and are written in different coding languages (in Norse mythology, Yggdrasil is an immense tree that connects many lands). Further efforts by the consortium will support researchers in visualizing model outputs to allow results to be intuitively interpreted. Consequently, multiscale models will steer research and technological advancements to better our understanding of crop responses to future climates, thus aiding global food security. (Summary by Caroline Dowling) Plant Journal 10.1111/tpj.14722
Throughout the plant science community, the use of computational or in silico analyses which precede traditional studies are gaining traction to identify research opportunities. Multiscale computational models are those which assimilate data from all biological system levels from gene to ecosystem. Benes et al. advocate for the use of such multiscale models to advise crop improvement studies. The authors present examples in which single scale mathematical models have previously guided research such as to inform crop management strategies. However, by combining models of different biological scales, we can evaluate crop “ideotypes” and test “what if” scenarios to comprehend how crops will react in untried conditions. To that end, we need to develop methods to bridge independently designed models, which is a major aim of the Crops in silico consortium. This group has developed the yggdrasil framework which facilitates connections between models that operate at distinct biological scales and are written in different coding languages (in Norse mythology, Yggdrasil is an immense tree that connects many lands). Further efforts by the consortium will support researchers in visualizing model outputs to allow results to be intuitively interpreted. Consequently, multiscale models will steer research and technological advancements to better our understanding of crop responses to future climates, thus aiding global food security. (Summary by Caroline Dowling) Plant Journal 10.1111/tpj.14722
Transient genome-wide interactions of the master transcription factor NLP7 initiate a rapid nitrogen-response cascade
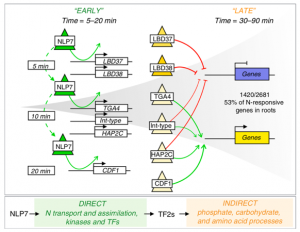 Transcription factors (TFs) and their genome-wide targets form gene regulatory networks that allow organisms to respond to stimuli. However, conventional biochemical assays only identify a subset of the TF-target interactions. In this paper, Alvarez et al. elucidate the genetic network of NIN-LIKE PROTEIN 7 (NLP7), which is a master regulator of plant nitrogen signaling. Both stable and transient NLP7 targets were identified by induction of NLP7 nuclear import coupled with time-series chromatin immunoprecipitation (ChIP-seq) and the use of a fusion to DNA adenine methyltransferase to mark transient interactions by DNA methylation. More than half (270/492) of the identified NLP7-target interactions are highly transient but changes in transcription continue even after NLP7 disassociates. These highly transient interactions are typically overlooked but crucial for early nitrogen response. The findings of this paper support the Hit-and-Run transcription model previously described by this group, in which transient interaction of a TF with its target sequence leads to long-term transcriptional changes. Moreover, the approaches to detect transient TF-target interactions on a genome-wide scale are applicable for other organisms. (Summary by Yun-Ting Kao) Nature Comms. 10.1038/s41467-020-14979-6
Transcription factors (TFs) and their genome-wide targets form gene regulatory networks that allow organisms to respond to stimuli. However, conventional biochemical assays only identify a subset of the TF-target interactions. In this paper, Alvarez et al. elucidate the genetic network of NIN-LIKE PROTEIN 7 (NLP7), which is a master regulator of plant nitrogen signaling. Both stable and transient NLP7 targets were identified by induction of NLP7 nuclear import coupled with time-series chromatin immunoprecipitation (ChIP-seq) and the use of a fusion to DNA adenine methyltransferase to mark transient interactions by DNA methylation. More than half (270/492) of the identified NLP7-target interactions are highly transient but changes in transcription continue even after NLP7 disassociates. These highly transient interactions are typically overlooked but crucial for early nitrogen response. The findings of this paper support the Hit-and-Run transcription model previously described by this group, in which transient interaction of a TF with its target sequence leads to long-term transcriptional changes. Moreover, the approaches to detect transient TF-target interactions on a genome-wide scale are applicable for other organisms. (Summary by Yun-Ting Kao) Nature Comms. 10.1038/s41467-020-14979-6
DRT111/SFPS splicing factor controls ABA sensitivity during seed development and germination
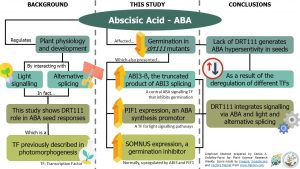 Abscisic acid (ABA) acts on different plant physiological and developmental processes by quite complex mechanisms. In fact, some of these processes interact with light signaling and some are controlled by alternative splicing. In this paper, Punzo et al. show that DRT111 – a splicing factor previously described in photomorphogenesis – regulates seed responses to ABA. The authors found that the germination of the two DRT11 knock-out mutants decreased after exposing their seeds to ABA, meaning the seeds were hypersensitive to ABA. To explain this phenotype, the authors analyzed the seed transcriptome of the drt111-2 mutant and found an up-regulated expression of PIF1 and SOMNUS, two transcription factors that inhibit germination. Interestingly, PIF1 is a transcription factor involved in light signaling pathways, showing that DRT111 affects the crosstalk between ABA and light signaling. Also, they found drt111-2 seeds accumulated ABI3-β –the non-functional, alternatively-spliced transcript of the germination inhibitor ABI3. Given this, the study shows that DRT111 also controls the alternative splicing of ABI3. (Summary by Carlos A. Ordóñez-Parra) Plant Physiol. 10.1104/pp.20.00037
Abscisic acid (ABA) acts on different plant physiological and developmental processes by quite complex mechanisms. In fact, some of these processes interact with light signaling and some are controlled by alternative splicing. In this paper, Punzo et al. show that DRT111 – a splicing factor previously described in photomorphogenesis – regulates seed responses to ABA. The authors found that the germination of the two DRT11 knock-out mutants decreased after exposing their seeds to ABA, meaning the seeds were hypersensitive to ABA. To explain this phenotype, the authors analyzed the seed transcriptome of the drt111-2 mutant and found an up-regulated expression of PIF1 and SOMNUS, two transcription factors that inhibit germination. Interestingly, PIF1 is a transcription factor involved in light signaling pathways, showing that DRT111 affects the crosstalk between ABA and light signaling. Also, they found drt111-2 seeds accumulated ABI3-β –the non-functional, alternatively-spliced transcript of the germination inhibitor ABI3. Given this, the study shows that DRT111 also controls the alternative splicing of ABI3. (Summary by Carlos A. Ordóñez-Parra) Plant Physiol. 10.1104/pp.20.00037
A molecular link between photoreceptors and jasmonates
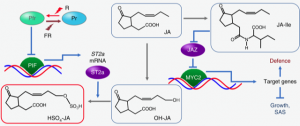 Plants balance their efforts on growth and defense depending on the situation. Competitive conditions signaled by the low ratios of red (R) to far-red (FR) light suppress defense responses mediated by the plant hormone jasmonate (JA), presumably for allocating more resources to promote plant growth. Fernández-Milmanda et al. identified the sulfotransferase ST2a as a missing link between photoreceptor-mediated light signaling and JA signaling. The authors found that low R:FR conditions induce ST2a in a manner dependent on the phyB-PIF pathway, a central regulator of light responses. ST2a catalyzes the sulfation of JA compounds to reduce the accumulation of bioactive JA conjugates. They demonstrated that ST2a is required for light-dependent signaling to mitigate JA responses and defense against pathogens. This study revealed a molecular mechanism by which light quality affects the homeostasis of JA compounds rewiring plant responses to the environment. (Summary by Tatsuya Nobori) Nature Plants 10.1038/s41477-020-0604-8
Plants balance their efforts on growth and defense depending on the situation. Competitive conditions signaled by the low ratios of red (R) to far-red (FR) light suppress defense responses mediated by the plant hormone jasmonate (JA), presumably for allocating more resources to promote plant growth. Fernández-Milmanda et al. identified the sulfotransferase ST2a as a missing link between photoreceptor-mediated light signaling and JA signaling. The authors found that low R:FR conditions induce ST2a in a manner dependent on the phyB-PIF pathway, a central regulator of light responses. ST2a catalyzes the sulfation of JA compounds to reduce the accumulation of bioactive JA conjugates. They demonstrated that ST2a is required for light-dependent signaling to mitigate JA responses and defense against pathogens. This study revealed a molecular mechanism by which light quality affects the homeostasis of JA compounds rewiring plant responses to the environment. (Summary by Tatsuya Nobori) Nature Plants 10.1038/s41477-020-0604-8
Integration of light signaling with endogenous developmental pathway to regulate flowering in Arabidopsis thaliana
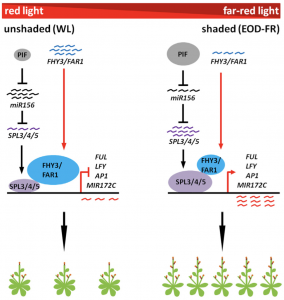 Response to changing environmental conditions is key for plant reproductive success. Previous studies have implicated FAR-RED ELONGATED HYPOCOTYL3 (FHY3) and FAR-RED IMPAIRED RESPONSE1 (FAR1) as inhibitors of flowering in phytochrome A signaling pathway. miR156-targeted SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) genes are involved in transition from juvenile to adult phase and promote flowering. In this paper Xie et al., showed that FHY3 and FAR1 protein levels decrease and that of SPL3/4/5 protein levels increase in response to shade, promoting flowering. Using biochemical and molecular genetic experiments they have shown that increased levels of SPLs activate the downstream target genes (FRUITFUL, LEAFY, APETALA1, MIR172C) by direct binding to the cis-regulatory elements in their promoters. The authors also show that under normal light conditions FHY3 and FAR1 interfere with SPLs by protein-protein interaction and this interaction weakens in shade, activating the downstream genes responsible for flowering. (Summary by Vijaya Batthula) Mol. Plant 10.1016/j.molp.2020.01.013
Response to changing environmental conditions is key for plant reproductive success. Previous studies have implicated FAR-RED ELONGATED HYPOCOTYL3 (FHY3) and FAR-RED IMPAIRED RESPONSE1 (FAR1) as inhibitors of flowering in phytochrome A signaling pathway. miR156-targeted SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) genes are involved in transition from juvenile to adult phase and promote flowering. In this paper Xie et al., showed that FHY3 and FAR1 protein levels decrease and that of SPL3/4/5 protein levels increase in response to shade, promoting flowering. Using biochemical and molecular genetic experiments they have shown that increased levels of SPLs activate the downstream target genes (FRUITFUL, LEAFY, APETALA1, MIR172C) by direct binding to the cis-regulatory elements in their promoters. The authors also show that under normal light conditions FHY3 and FAR1 interfere with SPLs by protein-protein interaction and this interaction weakens in shade, activating the downstream genes responsible for flowering. (Summary by Vijaya Batthula) Mol. Plant 10.1016/j.molp.2020.01.013
The M3Ks, a missing component discovered in the early ABA core signaling module
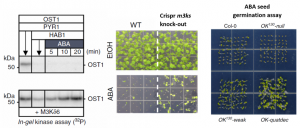 About a decade ago, the ABA receptors [PYR-ABACTIN RESISTANCE (PYR/PYL)/REGULATORY COMPONENT OF ABA RECEPTOR (RCAR)] were discovered, establishing the PYL-PP2C-SnRK2s core signalling module, in which ABA-binding to PYR/PYL receptors inhibits PP2C phosphatases, leading to activation of SnRK2 kinases, yet it remained unclear whether there are upstream protein kinases leading to SnRK2s activation via phosphorylation. Here, Takahashi et al. and Lin et al. have identified a family of MAP kinase kinase kinases (M3Ks), specifically the B2, B3 & B4 subfamilies of Raf-like protein kinases, that are required to (re)activate SnRK2s protein kinases to mediating osmotic and ABA responses. Takahashi and colleagues show that autophosphorylation of OST1 (SnRK2.6) is surprisingly insufficient for reactivation after dephosphorylation by PP2C, but the reactivation of OST1 could be rescued by M3Ks phosphorylation in vitro. The triple mutant of M3Ks (subgroup B3 members) has ABA and osmotic insensitive phenotypes in seed germination. Interestingly, this triple mutant shows stronger impairment in rapid osmotic stress activation of SnRK2 compared to ABA application, indicating that this M3Ks-SnRK2 cascade could also be required in ABA-independent osmotic stress signal transduction. Lin et al. generated higher order mutants of M3Ks subgroup B2/B3/B4, i.e., OK130-weak (B4 subgroup weak alleles), OK130-null (B4 subgroup mutant), OK-quatdec and -quindec mutants (B2, B4 and B4 subgroup mutant). OK130-null and OK-quatdec mutants are hypersensitive to osmotic stress and insensitive to ABA seed germination, respectively. Both studies uncover the M3Ks as the upstream kinases to trigger the SnRKs activation in ABA and osmotic stress responses. Further understanding this mechanism in crop plants will help to produce crops resistant to abiotic stresses. (Summary by Min May Wong) Nature Communication 10.1038/s41467-019-13875-y & 10.1038/s41467-020-14477-9
About a decade ago, the ABA receptors [PYR-ABACTIN RESISTANCE (PYR/PYL)/REGULATORY COMPONENT OF ABA RECEPTOR (RCAR)] were discovered, establishing the PYL-PP2C-SnRK2s core signalling module, in which ABA-binding to PYR/PYL receptors inhibits PP2C phosphatases, leading to activation of SnRK2 kinases, yet it remained unclear whether there are upstream protein kinases leading to SnRK2s activation via phosphorylation. Here, Takahashi et al. and Lin et al. have identified a family of MAP kinase kinase kinases (M3Ks), specifically the B2, B3 & B4 subfamilies of Raf-like protein kinases, that are required to (re)activate SnRK2s protein kinases to mediating osmotic and ABA responses. Takahashi and colleagues show that autophosphorylation of OST1 (SnRK2.6) is surprisingly insufficient for reactivation after dephosphorylation by PP2C, but the reactivation of OST1 could be rescued by M3Ks phosphorylation in vitro. The triple mutant of M3Ks (subgroup B3 members) has ABA and osmotic insensitive phenotypes in seed germination. Interestingly, this triple mutant shows stronger impairment in rapid osmotic stress activation of SnRK2 compared to ABA application, indicating that this M3Ks-SnRK2 cascade could also be required in ABA-independent osmotic stress signal transduction. Lin et al. generated higher order mutants of M3Ks subgroup B2/B3/B4, i.e., OK130-weak (B4 subgroup weak alleles), OK130-null (B4 subgroup mutant), OK-quatdec and -quindec mutants (B2, B4 and B4 subgroup mutant). OK130-null and OK-quatdec mutants are hypersensitive to osmotic stress and insensitive to ABA seed germination, respectively. Both studies uncover the M3Ks as the upstream kinases to trigger the SnRKs activation in ABA and osmotic stress responses. Further understanding this mechanism in crop plants will help to produce crops resistant to abiotic stresses. (Summary by Min May Wong) Nature Communication 10.1038/s41467-019-13875-y & 10.1038/s41467-020-14477-9
A feedforward loop controls vascular regeneration and tissue repair through local auxin biosynthesis
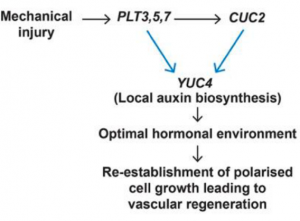 Plants are constantly exposed to biotic and biotic stresses that can cause tissue damage, and as a response plants have evolved remarkably plastic regenerative mechanisms in response to wounding. Although some genes required for regeneration have been identified in the Arabidopsis root context, most notably PLETHORA (PLT) genes, the molecular players required for wound repair in aerial organs are mostly unknown. Here, Radhakrishnan et al. revealed that PLT3, PLT5 and PLT7 (PLT3,5,7) transcription factor genes that are required for in vitro shoot regeneration, among other processes, as well as the closely related AINTEGUMENTA gene, are key players necessary for tissue repair and vascular regeneration in aerial organs. Through combinations of loss-of-function mutants, reporter and overexpression lines, the authors dissected a feedforward loop circuit that drives regeneration of vascular tissue in response to injury. The authors demonstrated that, in response to injury, PLT3,5,7 directly activate transcription of CUC2, a known regulator of PIN1 polarity and auxin distribution. The PLT-CUC2 module then synergistically activates transcription of the auxin biosynthesis gene YUC4. This feedforward loop creates an optimal hormonal environment that promotes tissue repair and vascular regeneration through PIN1 polarization and induction of vascular identity by AINTEGUMENTA. This study shows how stem cell regulators identified in other contexts can also serve as regulatory triggers in other developmental processes. (Summary by Jesus Leon) Development 10.1242/dev.185710
Plants are constantly exposed to biotic and biotic stresses that can cause tissue damage, and as a response plants have evolved remarkably plastic regenerative mechanisms in response to wounding. Although some genes required for regeneration have been identified in the Arabidopsis root context, most notably PLETHORA (PLT) genes, the molecular players required for wound repair in aerial organs are mostly unknown. Here, Radhakrishnan et al. revealed that PLT3, PLT5 and PLT7 (PLT3,5,7) transcription factor genes that are required for in vitro shoot regeneration, among other processes, as well as the closely related AINTEGUMENTA gene, are key players necessary for tissue repair and vascular regeneration in aerial organs. Through combinations of loss-of-function mutants, reporter and overexpression lines, the authors dissected a feedforward loop circuit that drives regeneration of vascular tissue in response to injury. The authors demonstrated that, in response to injury, PLT3,5,7 directly activate transcription of CUC2, a known regulator of PIN1 polarity and auxin distribution. The PLT-CUC2 module then synergistically activates transcription of the auxin biosynthesis gene YUC4. This feedforward loop creates an optimal hormonal environment that promotes tissue repair and vascular regeneration through PIN1 polarization and induction of vascular identity by AINTEGUMENTA. This study shows how stem cell regulators identified in other contexts can also serve as regulatory triggers in other developmental processes. (Summary by Jesus Leon) Development 10.1242/dev.185710
MicroRNAs and the control of stomatal development
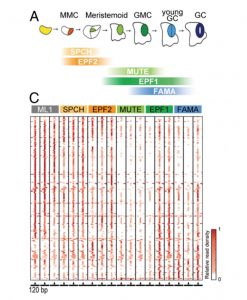 Stomata mediate critical functions in plant life: gas exchange, water loss, and some environmental responses. At the molecular level, some bHLH transcription factors and a MAP-kinase pathway control a series of asymmetric and symmetric cell divisions of stomatal stem cells to form a guard cell. In a recent study Zhu et al. explored the implication of microRNAs in the formation of stomata. They expressed AGO1 using the promoters of stage-specific marker genes (SPEECHLESS, MUTE, FAMA, EPF1, and EPF2) to isolate AGO1-miRNA complexes. They found that more than 60% of the annotated MIR genes in the Arabidopsis genome are dynamically expressed during stomatal development. They conducted in vivo analysis to test the functional relevance of some of them: the overexpression of miR829 and miR3932 altered stomatal development, generating an increase in the number of total stomata or stomata pairs. Further analyses included the prediction of potential mRNA targets of miRNAs; with these, authors uncovered cellular processes presumably controlled by the differentially expressed miRNAs during stomata formation. This study provides new insights about the dynamic control of stomatal development; for instance, the expression of miR399 in the stomata lineage is anticorrelated with the expression of PHO2, which regulates the PHO1 and PHT1 phosphate transporters, revealing a relationship between stomatal formation and nutrient homeostasis. (Summary by Humberto Herrera-Ubaldo) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1919722117
Stomata mediate critical functions in plant life: gas exchange, water loss, and some environmental responses. At the molecular level, some bHLH transcription factors and a MAP-kinase pathway control a series of asymmetric and symmetric cell divisions of stomatal stem cells to form a guard cell. In a recent study Zhu et al. explored the implication of microRNAs in the formation of stomata. They expressed AGO1 using the promoters of stage-specific marker genes (SPEECHLESS, MUTE, FAMA, EPF1, and EPF2) to isolate AGO1-miRNA complexes. They found that more than 60% of the annotated MIR genes in the Arabidopsis genome are dynamically expressed during stomatal development. They conducted in vivo analysis to test the functional relevance of some of them: the overexpression of miR829 and miR3932 altered stomatal development, generating an increase in the number of total stomata or stomata pairs. Further analyses included the prediction of potential mRNA targets of miRNAs; with these, authors uncovered cellular processes presumably controlled by the differentially expressed miRNAs during stomata formation. This study provides new insights about the dynamic control of stomatal development; for instance, the expression of miR399 in the stomata lineage is anticorrelated with the expression of PHO2, which regulates the PHO1 and PHT1 phosphate transporters, revealing a relationship between stomatal formation and nutrient homeostasis. (Summary by Humberto Herrera-Ubaldo) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1919722117
Convergent recruitment of TALE homeodomain life cycle regulators to direct sporophyte development in land plants and brown algae
 Life cycles in sexually reproducing plants and algae alternate between diploid (sporophytic) and haploid (gametophytic) generations. The haploid gametophyte produces gametes that mate to generate the diploid sporophyte, which in turn undergoes meiosis to generate haploid spores. Development must be coordinated with these life cycle-dependent changes in ploidy, so that mating and meiosis take place in the correct generation. How is this coordination achieved? Arun et al. studied this question in Ectocarpus, a brown alga well suited for genetic analysis of life cycle transitions. Normally, gamete fusion (mating) marks the beginning of the sporophyte generation. However, in Ectocarpus unmated gametes may also switch to the sporophytic fate. The haploid origin of these so-called “partheno-sporophytes” facilitates analysis of loss-of-function mutations in genes that act in the diploid sporophyte. Using this system, they identified two genes, OUROBOROS and SAMSARA, without which the would-be partheno-sporophyte develops into a fully functional gametophyte. Both OUR and SAM encode three amino acid extension homeodomain (TALE HD) transcription factors needed to turn on the sporophyte transcriptional program. Interestingly, the same TALE HD is found in transcription factors that activate the zygotic program in Chlamydomonas and sporophyte development in Physcomitrella, two green lineage organisms that are not closely related to brown algae. These finding point to a common, and ancient origin of the machinery controlling eukaryotic life cycle transitions. (Summary by Frej Tulin) 10.7554/eLife.43101
Life cycles in sexually reproducing plants and algae alternate between diploid (sporophytic) and haploid (gametophytic) generations. The haploid gametophyte produces gametes that mate to generate the diploid sporophyte, which in turn undergoes meiosis to generate haploid spores. Development must be coordinated with these life cycle-dependent changes in ploidy, so that mating and meiosis take place in the correct generation. How is this coordination achieved? Arun et al. studied this question in Ectocarpus, a brown alga well suited for genetic analysis of life cycle transitions. Normally, gamete fusion (mating) marks the beginning of the sporophyte generation. However, in Ectocarpus unmated gametes may also switch to the sporophytic fate. The haploid origin of these so-called “partheno-sporophytes” facilitates analysis of loss-of-function mutations in genes that act in the diploid sporophyte. Using this system, they identified two genes, OUROBOROS and SAMSARA, without which the would-be partheno-sporophyte develops into a fully functional gametophyte. Both OUR and SAM encode three amino acid extension homeodomain (TALE HD) transcription factors needed to turn on the sporophyte transcriptional program. Interestingly, the same TALE HD is found in transcription factors that activate the zygotic program in Chlamydomonas and sporophyte development in Physcomitrella, two green lineage organisms that are not closely related to brown algae. These finding point to a common, and ancient origin of the machinery controlling eukaryotic life cycle transitions. (Summary by Frej Tulin) 10.7554/eLife.43101
Formation of NB-LRR receptor ZAR1 resistome in vivo ($)
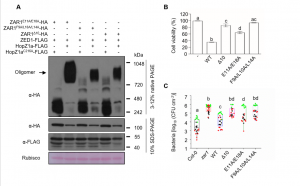 To fight against pathogenic microbes, plants have evolved intracellular nucleotide-binding, leucine-rich repeat (NB-LRR) receptors, known as NLRs, to monitor pathogen effector proteins and trigger robust immune responses. ZAR1 is a canonical NLR, and a forms a multimeric resistosome complex in vitro in association with partners and targets RKS1/PBL2/UMP/ATP. Now, Hu et al. have demonstrated that ZAR1 oligomerization is induced in vivo by the bacterial effectors AvrAC and HopZ1a. Moreover, the key residues for the in vivo HopZ1a-induced ZAR1 oligomerization and immunity are highly consistent with ZAR1-RKS1-PBL2UMP resistosome assembly in vitro. Furthermore, these same residues are essential for disease resistance, supporting a role for this complex in plant defense. (Summary by Nanxun Qin) Molecular Plant 10.1016/j.molp.2020.03.004
To fight against pathogenic microbes, plants have evolved intracellular nucleotide-binding, leucine-rich repeat (NB-LRR) receptors, known as NLRs, to monitor pathogen effector proteins and trigger robust immune responses. ZAR1 is a canonical NLR, and a forms a multimeric resistosome complex in vitro in association with partners and targets RKS1/PBL2/UMP/ATP. Now, Hu et al. have demonstrated that ZAR1 oligomerization is induced in vivo by the bacterial effectors AvrAC and HopZ1a. Moreover, the key residues for the in vivo HopZ1a-induced ZAR1 oligomerization and immunity are highly consistent with ZAR1-RKS1-PBL2UMP resistosome assembly in vitro. Furthermore, these same residues are essential for disease resistance, supporting a role for this complex in plant defense. (Summary by Nanxun Qin) Molecular Plant 10.1016/j.molp.2020.03.004
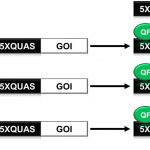
The Q-System as a synthetic transcriptional regulator in plants
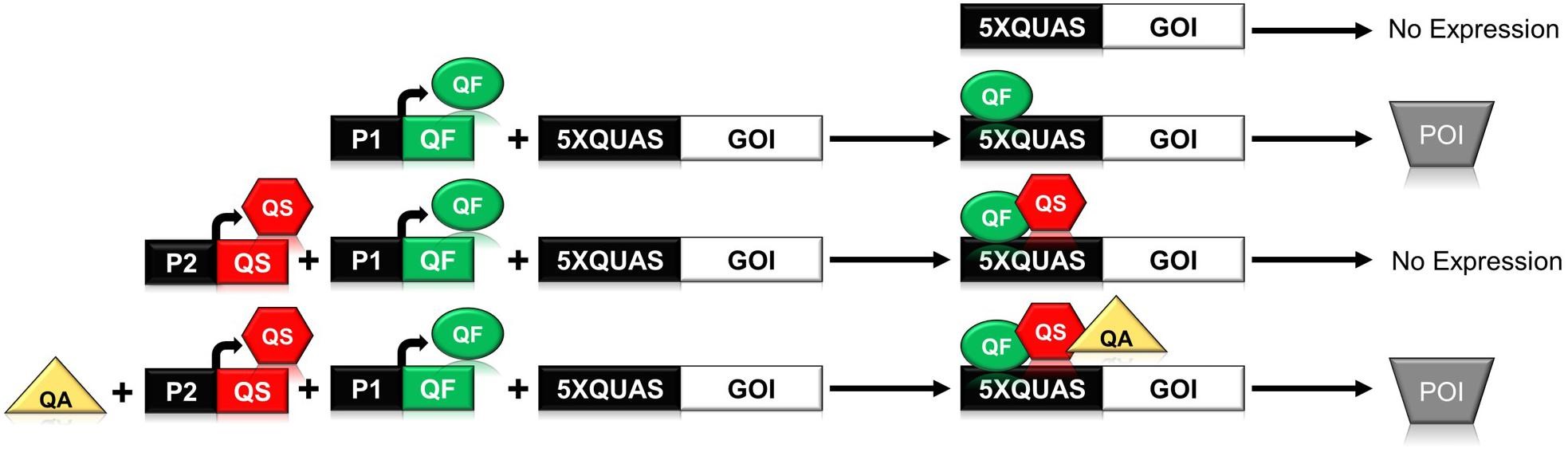 The ability to reliably induce a transgene has greatly enhanced the study of plant biology. Various chemical inducible system have worked robustly in plants, but plant synthetic biology is still lacking an efficient orthogonal (from outside) inducible system where multiple genes can be controlled at the same time. The Q-system from the fungus Neurospora crassa has been shown to be promising and constitutes a repressible binary system. This synthetic Q-system encodes both transcriptional activator (QF) and repressor (QS).The activation of a gene-of-interest (GOI) by QF can be inhibited by QS. Furthermore, addition of quinic acid (Qa) inhibits the activity of QS to reactivate the gene. In this paper, Persad et al. were able to validate the synthetic Q system in two different plant systems, soybean protoplasts and N. benthamiana leaves. The ability of the Q-system to coordinate the expression of multiple genes was determined through the expression of three fluorescent proteins from a single QF activator in a single cell. Overall, this paper has shown the potential of the Q system as a multidimensional tool kit in controlling and regulating transgene expression in plants. (Summary by Sunita Pathak) Frontiers in Plant Science 10.3389/fpls.2020.00245
The ability to reliably induce a transgene has greatly enhanced the study of plant biology. Various chemical inducible system have worked robustly in plants, but plant synthetic biology is still lacking an efficient orthogonal (from outside) inducible system where multiple genes can be controlled at the same time. The Q-system from the fungus Neurospora crassa has been shown to be promising and constitutes a repressible binary system. This synthetic Q-system encodes both transcriptional activator (QF) and repressor (QS).The activation of a gene-of-interest (GOI) by QF can be inhibited by QS. Furthermore, addition of quinic acid (Qa) inhibits the activity of QS to reactivate the gene. In this paper, Persad et al. were able to validate the synthetic Q system in two different plant systems, soybean protoplasts and N. benthamiana leaves. The ability of the Q-system to coordinate the expression of multiple genes was determined through the expression of three fluorescent proteins from a single QF activator in a single cell. Overall, this paper has shown the potential of the Q system as a multidimensional tool kit in controlling and regulating transgene expression in plants. (Summary by Sunita Pathak) Frontiers in Plant Science 10.3389/fpls.2020.00245