Plant Science Research Weekly: March 10, 2023
Review: Salt-tolerant crops: Time to deliver
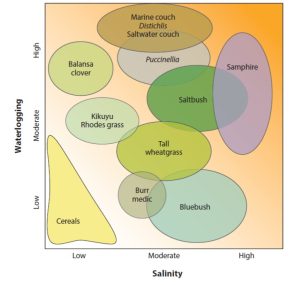 Few topics are as inherently interesting from both fundamental and applied perspectives as salt tolerance in crop plants. From the basic science side, cells have several strategies that they use to keep Na+ levels low in their cytosol in spite of what can be a very steep concentration gradient from out to in. Furthermore, at a physiological level, some tissues can cope with salt whereas others can’t, so plants use strategies to exclude out or sequester in salt. Finally, plants have a wide range of salinity tolerances, providing ample material to study. From the perspective of crop production, soil salinization is an increasing problem. Further, as the authors note, seawater incursion into agricultural regions is increasing with sea level rises and more vigorous storm systems. In this timely review, Melino and Tester survey our understanding of salt-tolerance in plants, with a particular focus on how to deliver salt-tolerant crops. They cover strategies from traditional breeding and selection, introgression of salinity tolerance from crop wild relatives or de novo domestication of these near crops, the use of genetic engineering to alter ion channels and membrane transporters, and a particularly interesting discussion of the use of grafting as a strategy to combine salt-tolerant root stocks with high-yielding shoot scions. They highlight a gap between some incentives for breeders (which include national yields) and small-scale farmers, particularly those whose lands are most affected. Although the challenges are significant, they end with the positive message that “together, we can do this!” (Summary by Mary Williams @PlantTeaching) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-061422-104322
Few topics are as inherently interesting from both fundamental and applied perspectives as salt tolerance in crop plants. From the basic science side, cells have several strategies that they use to keep Na+ levels low in their cytosol in spite of what can be a very steep concentration gradient from out to in. Furthermore, at a physiological level, some tissues can cope with salt whereas others can’t, so plants use strategies to exclude out or sequester in salt. Finally, plants have a wide range of salinity tolerances, providing ample material to study. From the perspective of crop production, soil salinization is an increasing problem. Further, as the authors note, seawater incursion into agricultural regions is increasing with sea level rises and more vigorous storm systems. In this timely review, Melino and Tester survey our understanding of salt-tolerance in plants, with a particular focus on how to deliver salt-tolerant crops. They cover strategies from traditional breeding and selection, introgression of salinity tolerance from crop wild relatives or de novo domestication of these near crops, the use of genetic engineering to alter ion channels and membrane transporters, and a particularly interesting discussion of the use of grafting as a strategy to combine salt-tolerant root stocks with high-yielding shoot scions. They highlight a gap between some incentives for breeders (which include national yields) and small-scale farmers, particularly those whose lands are most affected. Although the challenges are significant, they end with the positive message that “together, we can do this!” (Summary by Mary Williams @PlantTeaching) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-061422-104322
Review: Difficult situations cause blushing in plants, much as they do in people
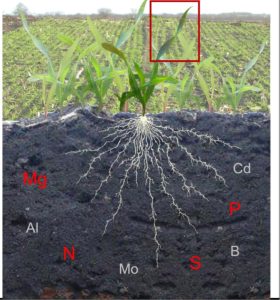 As with people, plants blush in difficult circumstances. Situations such as nutrient stress make plants uncomfortable, resulting in the accumulation of anthocyanins in different plant tissues and color changes in the tissues. Jezek et al. reviewed how and why anthocyanins accumulate in foliar tissues in nutrient-deficient plants. Anthocyanins accumulate in anthocyanoplasts or anthocyanic vacuoles in response to nutrient deficiencies, in patterns that are characteristic of the type of nutrient deficiency. N deficiency often results in chlorosis with leaf reddening or purpling in older leaves. P deviciency leads to increased chlorophyll density in the leaf accompanied by a dark blueish-green appearance, whereas Mg deficiency leads to chlorophyll degradation in mature leaves, and S deficiency causes purple coloration in young leaves. Color changes due to anthocyanin accumulation in micronutrient-deficient plants are also covered in this review. Other physiological responses resulting from anthocyanin accumulation under environmental stress are sugar accumulation, anthocyanins performing photoprotection roles, and ROS scavenging. Thus, anthocyanin-induced colour-changes in leaves can be used in designing optical devices for non-destructive diagnosis of nutrient stress in green plants. The genetic mechanisms underpinning anthocyanin accumulation in stressed conditions in various plants are still incompleteley understood. (Summary by Idowu Arinola Obisesan, @IdowuAobisesan) New Phytol. 10.1111/nph.18833
As with people, plants blush in difficult circumstances. Situations such as nutrient stress make plants uncomfortable, resulting in the accumulation of anthocyanins in different plant tissues and color changes in the tissues. Jezek et al. reviewed how and why anthocyanins accumulate in foliar tissues in nutrient-deficient plants. Anthocyanins accumulate in anthocyanoplasts or anthocyanic vacuoles in response to nutrient deficiencies, in patterns that are characteristic of the type of nutrient deficiency. N deficiency often results in chlorosis with leaf reddening or purpling in older leaves. P deviciency leads to increased chlorophyll density in the leaf accompanied by a dark blueish-green appearance, whereas Mg deficiency leads to chlorophyll degradation in mature leaves, and S deficiency causes purple coloration in young leaves. Color changes due to anthocyanin accumulation in micronutrient-deficient plants are also covered in this review. Other physiological responses resulting from anthocyanin accumulation under environmental stress are sugar accumulation, anthocyanins performing photoprotection roles, and ROS scavenging. Thus, anthocyanin-induced colour-changes in leaves can be used in designing optical devices for non-destructive diagnosis of nutrient stress in green plants. The genetic mechanisms underpinning anthocyanin accumulation in stressed conditions in various plants are still incompleteley understood. (Summary by Idowu Arinola Obisesan, @IdowuAobisesan) New Phytol. 10.1111/nph.18833
Opinion. Burning lignin: overlooked cues for post-fire seed germination
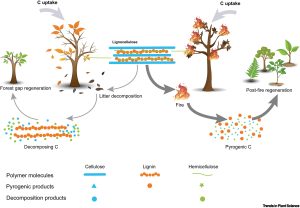 Smoke is a crucial germination trigger in ecosystems with naturally recurring fires, such as savannas and Mediterranean forests. There are various compounds in smoke, yet karrikins (KARs) –the primary product of cellulose combustion– have dominated the research efforts to elucidate the mechanisms behind smoke-promoted germination. In this piece, Cao and collaborators draw our attention to syringaldehyde (SAL), a product derived from lignin combustion with a recently described role in germination. While water can wash KARs away, SAL can be adsorbed and retained by soil particles and seeds. When the latter occurs, SAL is retained until proper germination conditions are met, acting as a “ready-to-use” germination cue. Such promoting effect has been shown in just a handful of species, so much more research is needed to evaluate the relative importance of such compounds and the evolution of such a response. However, the authors highlight that the origin of lignin and the expansion of vegetation dominated by vascular plants in the Paleozoic pretty well match the period of spreading fire event, a good scenario for the evolution of smoke-promoted germination mechanisms. Therefore, this piece points out an exciting research avenue that promises to provide a complete view of the mechanisms of smoke-promoted germination (Summary by Carlos A. Ordóñez-Parra, @caordonezparra) Trends Plant Sci. 10.1016/j.tplants.2023.01.011
Smoke is a crucial germination trigger in ecosystems with naturally recurring fires, such as savannas and Mediterranean forests. There are various compounds in smoke, yet karrikins (KARs) –the primary product of cellulose combustion– have dominated the research efforts to elucidate the mechanisms behind smoke-promoted germination. In this piece, Cao and collaborators draw our attention to syringaldehyde (SAL), a product derived from lignin combustion with a recently described role in germination. While water can wash KARs away, SAL can be adsorbed and retained by soil particles and seeds. When the latter occurs, SAL is retained until proper germination conditions are met, acting as a “ready-to-use” germination cue. Such promoting effect has been shown in just a handful of species, so much more research is needed to evaluate the relative importance of such compounds and the evolution of such a response. However, the authors highlight that the origin of lignin and the expansion of vegetation dominated by vascular plants in the Paleozoic pretty well match the period of spreading fire event, a good scenario for the evolution of smoke-promoted germination mechanisms. Therefore, this piece points out an exciting research avenue that promises to provide a complete view of the mechanisms of smoke-promoted germination (Summary by Carlos A. Ordóñez-Parra, @caordonezparra) Trends Plant Sci. 10.1016/j.tplants.2023.01.011
Review: Temperature sensing in plants
 Like all organisms, plants respond to changes in temperature by activating pathways that enable them to stay alive in spite of rising or lowering temperatures. Interestingly though, there is no universal temperature sensing mechanism across the domains of life. Changes in membrane fluidity (think of melting butter) are common sensors, as are rates of reactions (cooler = slower) or protein or RNA conformation (with added heat destabilizing hydrogen bonds). This excellent review by Kerbler and Wigge describes four well-characterized temperature response pathways, and evidence for various thermal sensors. The described pathways include the vernalization pathway (through which prolonged cold temperatures confers flowering competence), cold stress responses, heat shock responses, and thermomorphogenesis in which growth patterns are altered by elevated temperatures. The sensors or signal integrators discussed include several membrane-localized proteins that respond to changes in membrane fluidity, thermal reversion of phytochrome (the rate at which Pfr reverts back to Pr, which is highly temperature dependent), phase separation and biomolecular condensates, and the presence of the histone variant H2A.Z in nucleosomes. Temperature signaling through transcription, splicing, and protein turnover is also discussed. This interesting and accessible review also poses the question of whether studies on temperature sensing in plants can be leveraged to engineer greater thermal resilience into crop plants. Wouldn’t that be interesting! (Summary by Mary Williams @PlantTeaching) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-102820-102235
Like all organisms, plants respond to changes in temperature by activating pathways that enable them to stay alive in spite of rising or lowering temperatures. Interestingly though, there is no universal temperature sensing mechanism across the domains of life. Changes in membrane fluidity (think of melting butter) are common sensors, as are rates of reactions (cooler = slower) or protein or RNA conformation (with added heat destabilizing hydrogen bonds). This excellent review by Kerbler and Wigge describes four well-characterized temperature response pathways, and evidence for various thermal sensors. The described pathways include the vernalization pathway (through which prolonged cold temperatures confers flowering competence), cold stress responses, heat shock responses, and thermomorphogenesis in which growth patterns are altered by elevated temperatures. The sensors or signal integrators discussed include several membrane-localized proteins that respond to changes in membrane fluidity, thermal reversion of phytochrome (the rate at which Pfr reverts back to Pr, which is highly temperature dependent), phase separation and biomolecular condensates, and the presence of the histone variant H2A.Z in nucleosomes. Temperature signaling through transcription, splicing, and protein turnover is also discussed. This interesting and accessible review also poses the question of whether studies on temperature sensing in plants can be leveraged to engineer greater thermal resilience into crop plants. Wouldn’t that be interesting! (Summary by Mary Williams @PlantTeaching) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-102820-102235
Coordinated histone variant H2A.Z eviction and H3.3 deposition control plant thermomorphogenesis
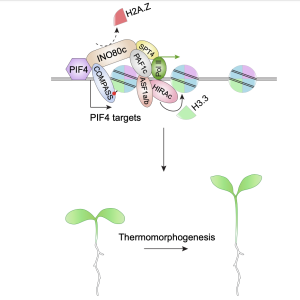 The effect of temperature on plant morphology is known as thermomorphogenesis. The phytohormone auxin is important for thermo-induced cell elongation. In Arabidopsis thaliana, Phytochrome-Interacting Factor 4 (PIF4) forms the central hub of the thermal response pathway and directly regulates auxin synthesis gene expression at high temperature. The activity of PIF4 is linked with chromatin regulation, and thus temperature influences epigenetic gene regulation in A. thaliana. PIF4, together with a chromatin remodeller INO80, is involved in H2A.Z eviction from the thermo-responsive gene loci in response to warm temperature. However, H2A.Z eviction isn’t sufficient for active transcription, and H2A.Z eviction is sometimes linked with active histone mark H3K4me3. Thus, H2A.Z eviction-induced gene regulation isn’t fully understood yet and further studies are essential for better understanding. Here, Zhao et al. showed that H3.3 deposition is required at warm temperature for thermomorphogenesis and RNA Pol II activity at PIF4 targets. The authors also showed that INO80 and ELF7 regulate H3.3 enrichment at PIF4 targets and that H3.3 is important for the H2A.Z eviction-induced gene activation. In a nutshell, Zhao et al. explain a striking association between H2A.Z eviction and H3.3 deposition leading to transcription activation at warm temperature and suggest a significant role of nucleosomal dynamics in the context of thermal response in A. thaliana. (Summary by Sourav Mukherjee @SouravBiotech) New Phytol. 10.1111/nph.18738
The effect of temperature on plant morphology is known as thermomorphogenesis. The phytohormone auxin is important for thermo-induced cell elongation. In Arabidopsis thaliana, Phytochrome-Interacting Factor 4 (PIF4) forms the central hub of the thermal response pathway and directly regulates auxin synthesis gene expression at high temperature. The activity of PIF4 is linked with chromatin regulation, and thus temperature influences epigenetic gene regulation in A. thaliana. PIF4, together with a chromatin remodeller INO80, is involved in H2A.Z eviction from the thermo-responsive gene loci in response to warm temperature. However, H2A.Z eviction isn’t sufficient for active transcription, and H2A.Z eviction is sometimes linked with active histone mark H3K4me3. Thus, H2A.Z eviction-induced gene regulation isn’t fully understood yet and further studies are essential for better understanding. Here, Zhao et al. showed that H3.3 deposition is required at warm temperature for thermomorphogenesis and RNA Pol II activity at PIF4 targets. The authors also showed that INO80 and ELF7 regulate H3.3 enrichment at PIF4 targets and that H3.3 is important for the H2A.Z eviction-induced gene activation. In a nutshell, Zhao et al. explain a striking association between H2A.Z eviction and H3.3 deposition leading to transcription activation at warm temperature and suggest a significant role of nucleosomal dynamics in the context of thermal response in A. thaliana. (Summary by Sourav Mukherjee @SouravBiotech) New Phytol. 10.1111/nph.18738
Single-residue substitution in histone H3 leads to over-lignification in Arabidopsis
 Chromatin changes are at the core of plant development and adaptation, as they constitute a crucial layer of gene expression control. The effects of different chromatin marks have historically been studied through mutations to the protein complexes depositing and removing the marks, which results in strong developmental defects and pleiotropic effects. Fal et al. expressed a histone H3 variant (H3.3) with a mutation in lysine 27 (lysine to alanine, or K27A) that prevents its modification. Arabidopsis plants expressing H3.3K27A displayed strong phenotypes, some of them resembling polycomb repressive complex 2 mutants (shorter stems, twisted leaves and early flowering) and some new ones, such as a marked thickening of the lignified tissue. A combination of transcriptomics and metabolomics analyses revealed that almost 70% of phenol-related metabolites mis-accumulated in H3.3K27A were phenylpropanoids, and that over 20% of genes involved in phenylpropanoid biosynthesis were differentially expressed in H3.3K27A. As phenylpropanoids are core components of lignin, the misregulation of their synthesis is likely to be linked to the observed over-lignification. Extending this strategy to additional histone residues could give new insights into the redundant and combinatorial nature of chromatin modifications. (Summary by Laura Turchi @turchi_l) New Phytol. 10.1111/nph.18666
Chromatin changes are at the core of plant development and adaptation, as they constitute a crucial layer of gene expression control. The effects of different chromatin marks have historically been studied through mutations to the protein complexes depositing and removing the marks, which results in strong developmental defects and pleiotropic effects. Fal et al. expressed a histone H3 variant (H3.3) with a mutation in lysine 27 (lysine to alanine, or K27A) that prevents its modification. Arabidopsis plants expressing H3.3K27A displayed strong phenotypes, some of them resembling polycomb repressive complex 2 mutants (shorter stems, twisted leaves and early flowering) and some new ones, such as a marked thickening of the lignified tissue. A combination of transcriptomics and metabolomics analyses revealed that almost 70% of phenol-related metabolites mis-accumulated in H3.3K27A were phenylpropanoids, and that over 20% of genes involved in phenylpropanoid biosynthesis were differentially expressed in H3.3K27A. As phenylpropanoids are core components of lignin, the misregulation of their synthesis is likely to be linked to the observed over-lignification. Extending this strategy to additional histone residues could give new insights into the redundant and combinatorial nature of chromatin modifications. (Summary by Laura Turchi @turchi_l) New Phytol. 10.1111/nph.18666
ETHYLENE INSENSITIVE3/EIN3-LIKE1 modulate FLOWERING LOCUS C expression via histone demethylase interaction
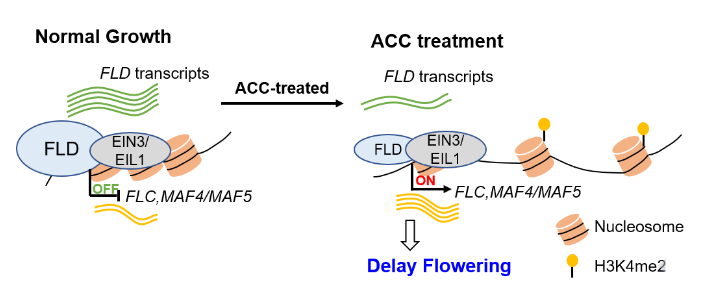 Flowering time is determined by both endogenous factors and environmental cues to ensure successful reproduction. ETHYLENE INSENSITIVE 3 (EIN3) and EIN3 LIKE 1 (EIL1) are transcription factors which are the key downstream regulators of ethylene signal transduction. Rapid flowering is promoted by the repression of FLOWERING LOCUS C (FLC). It has been seen that FLOWERING LOCUS D (FLD), which encodes for a demethylase H3K4me1/H3K4me2, binds to FLC loci, and removes methylation marks to repress it. Arabidopsis thaliana null mutants for EIN3 and EIL1 showed delayed flowering, however, the mechanism of this response is unknown. In this work, Xu et al., show how EIN3/EIL1 supress floral transition through the binding to the FLC loci and recruiting FLD. With a higher level of ethylene, FLD expression decreases which results in an hypermethylated H3K4me2 FLC locus, allowing its transcriptional activation and delaying flowering. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Physiol. 10.1093/plphys/kiad131
Flowering time is determined by both endogenous factors and environmental cues to ensure successful reproduction. ETHYLENE INSENSITIVE 3 (EIN3) and EIN3 LIKE 1 (EIL1) are transcription factors which are the key downstream regulators of ethylene signal transduction. Rapid flowering is promoted by the repression of FLOWERING LOCUS C (FLC). It has been seen that FLOWERING LOCUS D (FLD), which encodes for a demethylase H3K4me1/H3K4me2, binds to FLC loci, and removes methylation marks to repress it. Arabidopsis thaliana null mutants for EIN3 and EIL1 showed delayed flowering, however, the mechanism of this response is unknown. In this work, Xu et al., show how EIN3/EIL1 supress floral transition through the binding to the FLC loci and recruiting FLD. With a higher level of ethylene, FLD expression decreases which results in an hypermethylated H3K4me2 FLC locus, allowing its transcriptional activation and delaying flowering. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Physiol. 10.1093/plphys/kiad131
Pigment-regulating small siRNAs from YUP locus are responsible for speciation of monkeyflowers
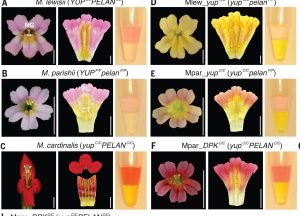 It has always been of interest to the plant community to understand the mechanisms behind the evolution of diverse floral carotenoid pigmentation within a genus. In this paper, Liang et al. investigated the mechanism behind monkeyflowers (Mimulus spp.) speciation driven by the flower color locus, YELLOW UPPER (YUP). YUP has been shown to control the presence or absence of yellow pigments in the petal, which subsequently affects pollinator choice and consequent reproductive speciation between closely related Mimulus species. RNA-seq data revealed a transcript with inverted repeats in the YUP locus which produced siRNAs in a phased pattern. The siRNAs were shown to concentrate in the petals and target the REDUCED CAROTENOID PIGMENTATION2 (RCP2) locus responsible for pigment accumulation in Mimulus. This extensive paper wrapped up with comparative genomics to understand the origin of YUP. Liang et al. presented another great example of the importance of noncoding genes and their role in flower speciation and evolution. (Summary by Apple Chew @_applechew) Science 10.1126/science.adg2774.
It has always been of interest to the plant community to understand the mechanisms behind the evolution of diverse floral carotenoid pigmentation within a genus. In this paper, Liang et al. investigated the mechanism behind monkeyflowers (Mimulus spp.) speciation driven by the flower color locus, YELLOW UPPER (YUP). YUP has been shown to control the presence or absence of yellow pigments in the petal, which subsequently affects pollinator choice and consequent reproductive speciation between closely related Mimulus species. RNA-seq data revealed a transcript with inverted repeats in the YUP locus which produced siRNAs in a phased pattern. The siRNAs were shown to concentrate in the petals and target the REDUCED CAROTENOID PIGMENTATION2 (RCP2) locus responsible for pigment accumulation in Mimulus. This extensive paper wrapped up with comparative genomics to understand the origin of YUP. Liang et al. presented another great example of the importance of noncoding genes and their role in flower speciation and evolution. (Summary by Apple Chew @_applechew) Science 10.1126/science.adg2774.
More warm-adapted species in soil seed banks than in herb layer plant communities across Europe
 Climate change profoundly impacts plant communities, for example, by changing the geographic range of some species and locally extinguishing others. Yet these changes take time, maybe a few years, to be observed. This lag between climate and plant communities changes could result from the soil seed bank (i.e., the reserve of viable, ungerminated seeds in and on the soil), which is assumed to buffer the effect of unsuitable conditions. In this exciting research, Auffret and collaborators use 54 soil seed bank datasets from studies across Western Europe to explore the role of the soil seed bank in mitigating the impacts of climate change. Species from warmer climates and broader distributions were found to produce seeds that persist more in the soil. In contrast, aboveground plant communities comprised species associated with relatively cooler temperatures. These results imply that soil seed banks might not delay but facilitate plant community changes towards communities dominated by warm-adapted species. Thus, species associated with cooler climates are more at risk of disappearing from local communities. As a result, this study provides fascinating insights into the mechanisms that control plant community responses to climate change (Summary by Carlos A. Ordóñez-Parra @caordonezparra) J. Ecol. 10.1111/1365-2745.14074
Climate change profoundly impacts plant communities, for example, by changing the geographic range of some species and locally extinguishing others. Yet these changes take time, maybe a few years, to be observed. This lag between climate and plant communities changes could result from the soil seed bank (i.e., the reserve of viable, ungerminated seeds in and on the soil), which is assumed to buffer the effect of unsuitable conditions. In this exciting research, Auffret and collaborators use 54 soil seed bank datasets from studies across Western Europe to explore the role of the soil seed bank in mitigating the impacts of climate change. Species from warmer climates and broader distributions were found to produce seeds that persist more in the soil. In contrast, aboveground plant communities comprised species associated with relatively cooler temperatures. These results imply that soil seed banks might not delay but facilitate plant community changes towards communities dominated by warm-adapted species. Thus, species associated with cooler climates are more at risk of disappearing from local communities. As a result, this study provides fascinating insights into the mechanisms that control plant community responses to climate change (Summary by Carlos A. Ordóñez-Parra @caordonezparra) J. Ecol. 10.1111/1365-2745.14074
Bioengineered “pikobodies” confer plant disease resistance
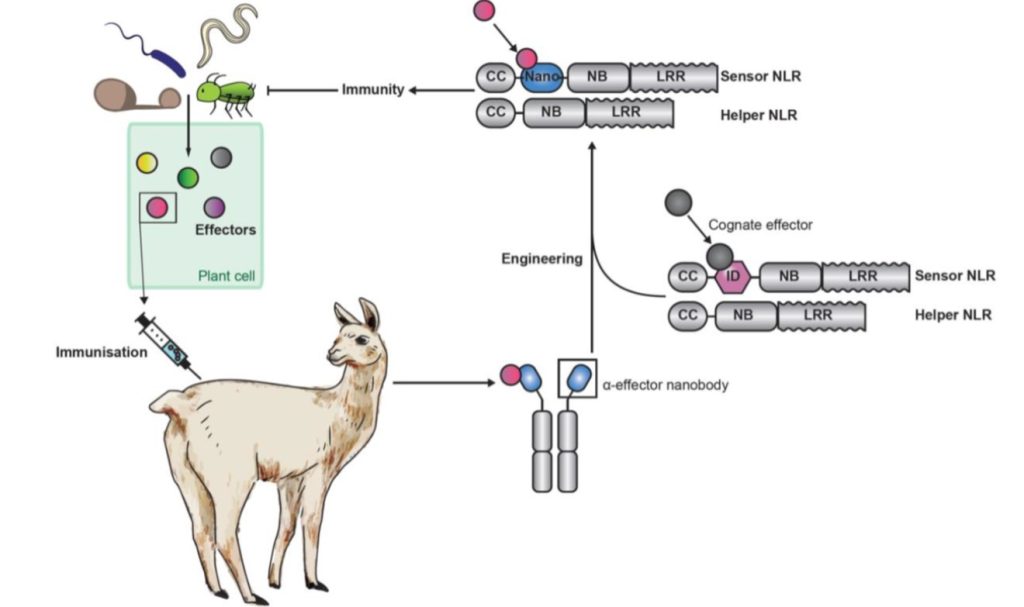 The vertebrate adaptive immune system is truly an evolutionary marvel. With its ability to mix-and-match segments of immunoglobulin genes, a nearly unlimited diversity of antigens can be recognized. Plants lack this ability, greatly limiting the number of antigens (and pathogens) any individual can recognize. Kourelis et al. did a “what if” experiment; what if plants could acquire some features of this adaptive immune system? To address this, they created fusions of segments of Pik-1, a NLR plant immune protein, to variable domains of naturally occurring single-chain antibodies that are produced in camelid animals and that are known as nanobodies. The results of this fusion combine the versatility of the animal antigen recognition domain with the endogenous immune function of the plant protein. As proof of concept, they spliced nanobody domains recognizing GFP or RFP into Pik-1, introduced the new protein along with its activity partner Pik-2 (they pair is collectively referred to as a pikobody) in Nicotiana benthamiana leaves and showed that the pikobodies promote an immune response in the presence of the corresponding fluorescent protein; in effect, they turned the fluorescent proteins into synthetic avirulence proteins (AVRs). The authors further showed that the pikobodies confer resistance to viruses carrying the fluorescent-protein antigen. As they observe, this technology could lead to “made to order” resistance genes; a pathogen effector would be used to raise nanobodies in an animal, with the corresponding nanobody gene lending its recognition domain to the pikobodies, which would be expressed in the plant. And bingo! Pathogen recognized and defenses deployed. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.abn4116
The vertebrate adaptive immune system is truly an evolutionary marvel. With its ability to mix-and-match segments of immunoglobulin genes, a nearly unlimited diversity of antigens can be recognized. Plants lack this ability, greatly limiting the number of antigens (and pathogens) any individual can recognize. Kourelis et al. did a “what if” experiment; what if plants could acquire some features of this adaptive immune system? To address this, they created fusions of segments of Pik-1, a NLR plant immune protein, to variable domains of naturally occurring single-chain antibodies that are produced in camelid animals and that are known as nanobodies. The results of this fusion combine the versatility of the animal antigen recognition domain with the endogenous immune function of the plant protein. As proof of concept, they spliced nanobody domains recognizing GFP or RFP into Pik-1, introduced the new protein along with its activity partner Pik-2 (they pair is collectively referred to as a pikobody) in Nicotiana benthamiana leaves and showed that the pikobodies promote an immune response in the presence of the corresponding fluorescent protein; in effect, they turned the fluorescent proteins into synthetic avirulence proteins (AVRs). The authors further showed that the pikobodies confer resistance to viruses carrying the fluorescent-protein antigen. As they observe, this technology could lead to “made to order” resistance genes; a pathogen effector would be used to raise nanobodies in an animal, with the corresponding nanobody gene lending its recognition domain to the pikobodies, which would be expressed in the plant. And bingo! Pathogen recognized and defenses deployed. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.abn4116



