Plant Science Research Weekly: July 26th
Review: Sulfated plant peptide hormones
 It’s hard to believe that when I was a student we were taught that “plants don’t have peptide hormones”. Since then we’ve discovered many diverse families of plant peptide hormones (see the Teaching Tool on peptide hormones for an excellent overview). Here, Kaufmann and Sauter review one family, the sulfated peptide hormones. Tyrosine sulfation is irreversible, lends specificity to peptide/receptor interactions, links the hormone’s activity to sulfur metabolism, and requires a plant-specific tyrosylprotein sulfotransferase. There are several types of sulfated peptide hormones that have been identified, including RGFs/GLVs/CLELs (root developmental responses), PSK and PSYs (cell expansion and damage responses), and CIFs (Casparian strip formation). This review updates the functions of each, what is known about their receptors and signaling pathways, and their potential applications. (Summary by Mary Williams) J. Exp. Bot. 10.1093/jxb/erz292
It’s hard to believe that when I was a student we were taught that “plants don’t have peptide hormones”. Since then we’ve discovered many diverse families of plant peptide hormones (see the Teaching Tool on peptide hormones for an excellent overview). Here, Kaufmann and Sauter review one family, the sulfated peptide hormones. Tyrosine sulfation is irreversible, lends specificity to peptide/receptor interactions, links the hormone’s activity to sulfur metabolism, and requires a plant-specific tyrosylprotein sulfotransferase. There are several types of sulfated peptide hormones that have been identified, including RGFs/GLVs/CLELs (root developmental responses), PSK and PSYs (cell expansion and damage responses), and CIFs (Casparian strip formation). This review updates the functions of each, what is known about their receptors and signaling pathways, and their potential applications. (Summary by Mary Williams) J. Exp. Bot. 10.1093/jxb/erz292
Perspective: Grazing animals drove domestication of grain crops
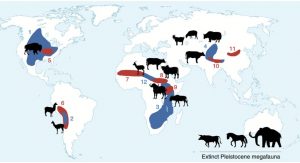 Here’s an interesting question: Without human intervention, why would one find a dense stand of plants, growing in rather nutrient-rich soil? Perhaps you recognized that these conditions suggest seed dispersal by endozoochory, which involves passage through an animal’s digestive tract. Spengler and Mueller suggest that human domestication of some grain crops may have started from such humble roots. They demonstrate that many domesticated grains have shifted from traits that make them survive passage through an animal’s gut to traits that make them easier for humans to harvest and store. This is an intriguing Perspective, sure to provide food for thought especially for students. (Summary by Mary Williams) Nature Plants 10.1038/s41477-019-0470-4
Here’s an interesting question: Without human intervention, why would one find a dense stand of plants, growing in rather nutrient-rich soil? Perhaps you recognized that these conditions suggest seed dispersal by endozoochory, which involves passage through an animal’s digestive tract. Spengler and Mueller suggest that human domestication of some grain crops may have started from such humble roots. They demonstrate that many domesticated grains have shifted from traits that make them survive passage through an animal’s gut to traits that make them easier for humans to harvest and store. This is an intriguing Perspective, sure to provide food for thought especially for students. (Summary by Mary Williams) Nature Plants 10.1038/s41477-019-0470-4
COP1 can be hijacked by photoreceptors via their VP motifs in Arabidopsis
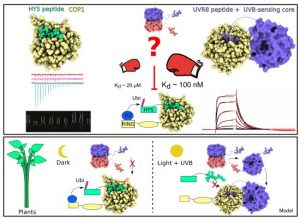 Plant development is characterized by a high degree of plasticity in response to light. Light‐activated plant photoreceptors bind and inhibit the E3 ubiquitin ligase COP1, thus protecting downstream transcription factors from degradation. However, the detailed mechanisms of how COP1 can function between upstream photoreceptors and downstream targets is enigmatic. Recently Lau et al. found that the activated UVR8 and cryptochrome photoreceptors and their signaling components compete for COP1. This process is mediated by a conserved VP peptide motif on them, allowing photoreceptors to displace the downstream targets from COP1 in a light-dependent manner. This VP peptide-based competition suggests that COP1 may utilize sophisticated mechanisms to coordinately regulate the transcription of specific genes under different type of light. (Summarized by Nanxun Qin) EMBO J. 10.15252/embj.2019102140
Plant development is characterized by a high degree of plasticity in response to light. Light‐activated plant photoreceptors bind and inhibit the E3 ubiquitin ligase COP1, thus protecting downstream transcription factors from degradation. However, the detailed mechanisms of how COP1 can function between upstream photoreceptors and downstream targets is enigmatic. Recently Lau et al. found that the activated UVR8 and cryptochrome photoreceptors and their signaling components compete for COP1. This process is mediated by a conserved VP peptide motif on them, allowing photoreceptors to displace the downstream targets from COP1 in a light-dependent manner. This VP peptide-based competition suggests that COP1 may utilize sophisticated mechanisms to coordinately regulate the transcription of specific genes under different type of light. (Summarized by Nanxun Qin) EMBO J. 10.15252/embj.2019102140
SPX4 acts on PHR1-dependent and -independent regulation of shoot phosphorus status
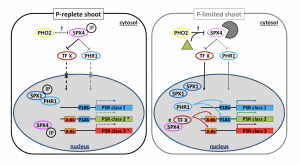 Plant phosphorus homeostasis is important to understand as humans seek to increase food production while also reducing the amount of agricultural inputs and environmental pollution. In their recent paper, Osorio and colleagues expand our current understanding of phosphorus homeostasis in Arabidopsis thaliana through their focus on SPX4 regulation of the phosphorus starvation response. SPX4 is one of a four-member family of negative regulators of phosphorus uptake in Arabidopsis. The authors identified that SPX4 abundance is controlled by the rate of protein turnover based on internal phosphorus concentration. However, unlike rice OsSPX4, this turnover isn’t based on common proteasomal degradation, nor is it responsive to external phosphite (Phi) supply. SPX4 also showed phosphorus-dependent sequestration of PHR1, an important phosphorus starvation response transcription factor, in the cytosol. Furthermore, the use of spx4 mutants demonstrated that misregulation of many the phosphorus starvation response genes was dependent on SPX4, PHR1, and PHO2. (Summary by Nathan Scinto-Madonich) Plant Physiol.10.1104/pp.18.00594
Plant phosphorus homeostasis is important to understand as humans seek to increase food production while also reducing the amount of agricultural inputs and environmental pollution. In their recent paper, Osorio and colleagues expand our current understanding of phosphorus homeostasis in Arabidopsis thaliana through their focus on SPX4 regulation of the phosphorus starvation response. SPX4 is one of a four-member family of negative regulators of phosphorus uptake in Arabidopsis. The authors identified that SPX4 abundance is controlled by the rate of protein turnover based on internal phosphorus concentration. However, unlike rice OsSPX4, this turnover isn’t based on common proteasomal degradation, nor is it responsive to external phosphite (Phi) supply. SPX4 also showed phosphorus-dependent sequestration of PHR1, an important phosphorus starvation response transcription factor, in the cytosol. Furthermore, the use of spx4 mutants demonstrated that misregulation of many the phosphorus starvation response genes was dependent on SPX4, PHR1, and PHO2. (Summary by Nathan Scinto-Madonich) Plant Physiol.10.1104/pp.18.00594
Plasma membrane-associated receptor like kinases relocalize to plasmodesmata in response to osmotic stress
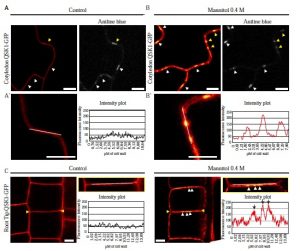 Plasmodesmata are channels through the cell wall that allow molecules and substances to move back and forth as needed; they also play a central role in growth, development and defence of all higher plants. In this study, Grison et al. describe the rapid relocation to the plasmodesmata pores of two plasma membrane-located Leucine-Rich-Repeat Receptor-Like-Kinases (LRR-RLKs), QSK1 and IMK2, in response to osmotic stress; they also show that this is not a general feature of LRR-RLKs. In particular, exposure to NaCl or mannitol triggers the relocalization in a QSK1-phosphorylation dependent manner within less than 2 min. Plasmodesmata aperture is regulated in part by callose deposition and removal. Lateral root (LR) development is also associated with callose-mediated plasmodesmata regulation, and the authors show here that the callose levels in the qsk1 mutant are reduced. The authors conclude that QSK1 has an important function not only in LR development but is required also to regulate callose deposition in response to mannitol. (Summarized by Francesca Resentini) Plant Physiol. 10.1104/pp.19.00473
Plasmodesmata are channels through the cell wall that allow molecules and substances to move back and forth as needed; they also play a central role in growth, development and defence of all higher plants. In this study, Grison et al. describe the rapid relocation to the plasmodesmata pores of two plasma membrane-located Leucine-Rich-Repeat Receptor-Like-Kinases (LRR-RLKs), QSK1 and IMK2, in response to osmotic stress; they also show that this is not a general feature of LRR-RLKs. In particular, exposure to NaCl or mannitol triggers the relocalization in a QSK1-phosphorylation dependent manner within less than 2 min. Plasmodesmata aperture is regulated in part by callose deposition and removal. Lateral root (LR) development is also associated with callose-mediated plasmodesmata regulation, and the authors show here that the callose levels in the qsk1 mutant are reduced. The authors conclude that QSK1 has an important function not only in LR development but is required also to regulate callose deposition in response to mannitol. (Summarized by Francesca Resentini) Plant Physiol. 10.1104/pp.19.00473
A calmodulin-gated calcium channel links pathogen patterns to plant immunity ($)
 Calcium ions mediate calcium-based defense responses in pattern triggered immunity (PTI) upon detection of pathogen patterns by plant surface receptors. A new study by Tian et al. has elucidated the molecular events that activate the calcium response. The authors show that adequeate calcium nutrient status is required to trigger immune response. EMS mutagenesis screeing identified two genes that encode for cyclic nucleotide-gated channel (CNGC) proteins, CNGC2 and CNGC4, which are essential for flg22 induced calcium burst. A reconstitution experiment using CNGC2 and CNGC4 in Xenopus oocytes produced a Ca2+ influx suggesting that the two partners form an active channel in Ca2+-mediated immune response. Furthermore, the authors showed that the PTI effector kinase BIK1 directly interacts with the formed channel and activates the channel possibly by the identified phospho-sites on the BIK1 protein. Hence this work illuminates a crucial missing link between the pattern-recognition receptor complex and calcium-dependent immunity during plant pathogenesis. (Summary by Amey Redkar) Nature. 10.1038/s41586-019-1413-y
Calcium ions mediate calcium-based defense responses in pattern triggered immunity (PTI) upon detection of pathogen patterns by plant surface receptors. A new study by Tian et al. has elucidated the molecular events that activate the calcium response. The authors show that adequeate calcium nutrient status is required to trigger immune response. EMS mutagenesis screeing identified two genes that encode for cyclic nucleotide-gated channel (CNGC) proteins, CNGC2 and CNGC4, which are essential for flg22 induced calcium burst. A reconstitution experiment using CNGC2 and CNGC4 in Xenopus oocytes produced a Ca2+ influx suggesting that the two partners form an active channel in Ca2+-mediated immune response. Furthermore, the authors showed that the PTI effector kinase BIK1 directly interacts with the formed channel and activates the channel possibly by the identified phospho-sites on the BIK1 protein. Hence this work illuminates a crucial missing link between the pattern-recognition receptor complex and calcium-dependent immunity during plant pathogenesis. (Summary by Amey Redkar) Nature. 10.1038/s41586-019-1413-y
Reconstituting an NLR cell death branch
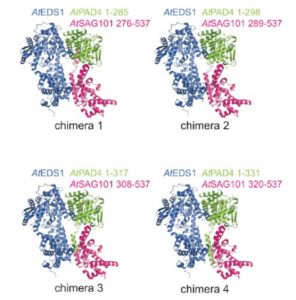 Arabidopsis genetics has identified numerous genes required for pathogen perception and defense response activation. Some of these genes are functionally conserved across plants and others are not. Toll-Interleukin1-Receptor (TIR)-domain NLRs (TNLs) are immune receptors that function upstream of EDS1 (enhanced disease susceptibility) genes. Lapin, Kovacova, Sun, et al. set out to investigate the phylogeny and function of EDS1 family members; these include EDS1, PAD4 and SAG101. In Arabidopsis, previous studies showed that AtEDS1 functions in a heterodimer with one of its partners, AtPAD4, to transcriptionally mobilize anti-microbial defense pathways. In this new work, the authors show that AtEDS1 and AtSAG101, together with AtNRG1, promote cell death in Arabidopsis. Previous work found that SAG101 is missing from monocot genomes, and here the authors also found it missing from the genomes of several eudicots. Through domain swapping and functional assays in Nicotiana benthamiana, the authors identified key domains required for heterodimerization and function in these related proteins. The authors conclude, “TNL-triggered cell death and pathogen growth restriction are determined by distinctive features of EDS1-SAG101 and EDS1-PAD4 complexes and that these signaling machineries coevolved with other components within plant species or clades to regulate downstream pathways in TNL immunity.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00118
Arabidopsis genetics has identified numerous genes required for pathogen perception and defense response activation. Some of these genes are functionally conserved across plants and others are not. Toll-Interleukin1-Receptor (TIR)-domain NLRs (TNLs) are immune receptors that function upstream of EDS1 (enhanced disease susceptibility) genes. Lapin, Kovacova, Sun, et al. set out to investigate the phylogeny and function of EDS1 family members; these include EDS1, PAD4 and SAG101. In Arabidopsis, previous studies showed that AtEDS1 functions in a heterodimer with one of its partners, AtPAD4, to transcriptionally mobilize anti-microbial defense pathways. In this new work, the authors show that AtEDS1 and AtSAG101, together with AtNRG1, promote cell death in Arabidopsis. Previous work found that SAG101 is missing from monocot genomes, and here the authors also found it missing from the genomes of several eudicots. Through domain swapping and functional assays in Nicotiana benthamiana, the authors identified key domains required for heterodimerization and function in these related proteins. The authors conclude, “TNL-triggered cell death and pathogen growth restriction are determined by distinctive features of EDS1-SAG101 and EDS1-PAD4 complexes and that these signaling machineries coevolved with other components within plant species or clades to regulate downstream pathways in TNL immunity.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00118
An N- terminal motif in NLR immune receptors is functionally conserved across distantly related plant species
 Plants have robust NLR (nucleotide-binding, leucine-rich repeat proteins) immune signaling networks consisting of sensor NLRs and helper NLRs that counteract diverse plant pathogens by inducing cell death at sites of pathogen infection. A recent preprint by Adachi et al. has defined an N terminal motif (MADA motif) that contributes to NLR-mediated cell death. By using in vitro Mu transposition insertion mutagenesis, the authors revealed a 29 amino acid region in the helper NLR NRC4 that on its own induces cell death. This motif is 50% identical in NLR ZAR1 and is also in 20% of the CC-NLRs found across dicot and monocot species. MADA swapping experiments have shown that the ZAR1 a1 helix and the N-termini of other MADA-CC-NLRs can not only functionally complement NRC4-mediated cell death activation but can also confer resistance to pathogens. Hence, this work provides new avenues to understand plant immune activation and resistance signalling, which ultimately can be harnessed to curtail pathogen-associated crop loss. (Summary by Amey Redkar) bioRxiv. 10.1101/693291
Plants have robust NLR (nucleotide-binding, leucine-rich repeat proteins) immune signaling networks consisting of sensor NLRs and helper NLRs that counteract diverse plant pathogens by inducing cell death at sites of pathogen infection. A recent preprint by Adachi et al. has defined an N terminal motif (MADA motif) that contributes to NLR-mediated cell death. By using in vitro Mu transposition insertion mutagenesis, the authors revealed a 29 amino acid region in the helper NLR NRC4 that on its own induces cell death. This motif is 50% identical in NLR ZAR1 and is also in 20% of the CC-NLRs found across dicot and monocot species. MADA swapping experiments have shown that the ZAR1 a1 helix and the N-termini of other MADA-CC-NLRs can not only functionally complement NRC4-mediated cell death activation but can also confer resistance to pathogens. Hence, this work provides new avenues to understand plant immune activation and resistance signalling, which ultimately can be harnessed to curtail pathogen-associated crop loss. (Summary by Amey Redkar) bioRxiv. 10.1101/693291
Root development is maintained by specific bacteria-bacteria interactions
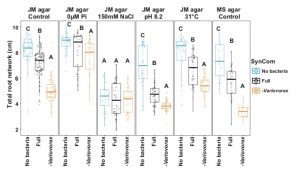 Understanding plant-microbe and microbe-microbe interactions is challening. Both kinds of interactions have significant impacts on plant health and nutritional uptake. Finkel et al. address how microbe-microbe interactions shape plant phenotypes by using the synthetic microbial community (SynCom) consisting of 185 bacterial members. Plants and SynCom were exposed to variable abiotic conditions such as salinity, temperature, pH and phosphate concentration. By analyzing the microbial community structure by 16S rRNA amplicon sequencing, the authors found that members of the bacterial genus Variovorax have a strong capacity to suppress root growth inhibition (RGI) by other bacteria members. This suppression was found to be robust against wide variety of RGI inducers, and was achieved by interfering in auxin and/or ethylene signaling. (Summary by Mugdha Sabale) bioRxiv 10.1101/645655
Understanding plant-microbe and microbe-microbe interactions is challening. Both kinds of interactions have significant impacts on plant health and nutritional uptake. Finkel et al. address how microbe-microbe interactions shape plant phenotypes by using the synthetic microbial community (SynCom) consisting of 185 bacterial members. Plants and SynCom were exposed to variable abiotic conditions such as salinity, temperature, pH and phosphate concentration. By analyzing the microbial community structure by 16S rRNA amplicon sequencing, the authors found that members of the bacterial genus Variovorax have a strong capacity to suppress root growth inhibition (RGI) by other bacteria members. This suppression was found to be robust against wide variety of RGI inducers, and was achieved by interfering in auxin and/or ethylene signaling. (Summary by Mugdha Sabale) bioRxiv 10.1101/645655
Compositional analysis of genetically engineered GR2E “Golden Rice”
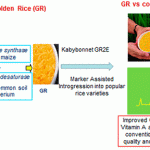
 In Southeast Asian countries, rice accounts for two-thirds of the caloric intake for many people. This narrow dietary base is often correlated with vitamin A deficiency, which can cause blindness, anemia and weakened immunity. In an effort to enhance vitamin A consumption, rice has been engineered to produce provitamin A carotenoids in the grain; this causes the grain to appear yellow, so the resulting rice is referred to as “Golden Rice”. Early versions using a phytoene synthase (psy) from daffodil have been improved upon through the use of a psy gene from maize, producing GR2E. Here, Swamy et al. have analyzed the composition of GR2E grown at four locations in the Philippines during 2015 and 2016 and found, “The only biologically meaningful difference between GR2E and control rice was in levels of β-carotene and other provitamin A carotenoids in the grain,” with the levels in the grain sufficient to significantly enhance human nutrition. The nutritionally-enhanced rice continues to move slowly through regulatory hurdles, but there is hope that it will soon be made available to consumers. (Summary by Mary Williams) J. Agric. Food Chem. 10.1021/acs.jafc.9b01524
In Southeast Asian countries, rice accounts for two-thirds of the caloric intake for many people. This narrow dietary base is often correlated with vitamin A deficiency, which can cause blindness, anemia and weakened immunity. In an effort to enhance vitamin A consumption, rice has been engineered to produce provitamin A carotenoids in the grain; this causes the grain to appear yellow, so the resulting rice is referred to as “Golden Rice”. Early versions using a phytoene synthase (psy) from daffodil have been improved upon through the use of a psy gene from maize, producing GR2E. Here, Swamy et al. have analyzed the composition of GR2E grown at four locations in the Philippines during 2015 and 2016 and found, “The only biologically meaningful difference between GR2E and control rice was in levels of β-carotene and other provitamin A carotenoids in the grain,” with the levels in the grain sufficient to significantly enhance human nutrition. The nutritionally-enhanced rice continues to move slowly through regulatory hurdles, but there is hope that it will soon be made available to consumers. (Summary by Mary Williams) J. Agric. Food Chem. 10.1021/acs.jafc.9b01524