Plant Science Research Weekly: January 7, 2022
Happy New Year! Welcome to our latest edition of Plant Science Research Weekly, featuring contributions from several of our new cohort of Plantae Fellows, who will be helping us keep up with exciting developments in plant sciences. Enjoy!
The evolutionary advantage of losing genes after endosymbiosis
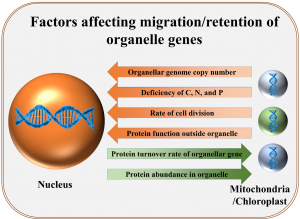 The emergence of eukaryotes is associated with endosymbiosis of mitochondria and chloroplast. In the course of evolution, most of the genes from these organelles have either migrated to the nucleus or been lost. But why have a small but significant number of genes remained in these organelle genomes? A recent analysis by Steven Kelly sheds light on possible reasons behind this. The study emphasizes that since organelles are present in large numbers in each cell, a relatively large amount of energy (ATP) is required to replicate their DNA. On the other hand, it is less expensive for a cell to maintain a gene in the nucleus and export the encoded protein to the organelles, especially if the protein is present in low abundance. The genes that are retained in these organelles are mainly the ones that encode proteins present at high levels, making their production within the organelles more economical. Other factors favoring gene migration to the nucleus include: additional functions of the gene product outside the organelle, nutritional deficiency of carbon, nitrogen, and phosphorus (constituents of DNA), and a low rate of protein turnover. (Summary and modified image by Kamal Kumar Malukani, @KamalMalukani). Genome Biol. 10.1186/s13059-021-02567-w
The emergence of eukaryotes is associated with endosymbiosis of mitochondria and chloroplast. In the course of evolution, most of the genes from these organelles have either migrated to the nucleus or been lost. But why have a small but significant number of genes remained in these organelle genomes? A recent analysis by Steven Kelly sheds light on possible reasons behind this. The study emphasizes that since organelles are present in large numbers in each cell, a relatively large amount of energy (ATP) is required to replicate their DNA. On the other hand, it is less expensive for a cell to maintain a gene in the nucleus and export the encoded protein to the organelles, especially if the protein is present in low abundance. The genes that are retained in these organelles are mainly the ones that encode proteins present at high levels, making their production within the organelles more economical. Other factors favoring gene migration to the nucleus include: additional functions of the gene product outside the organelle, nutritional deficiency of carbon, nitrogen, and phosphorus (constituents of DNA), and a low rate of protein turnover. (Summary and modified image by Kamal Kumar Malukani, @KamalMalukani). Genome Biol. 10.1186/s13059-021-02567-w
Sheathing leaf: The grass leaf conundrum!!
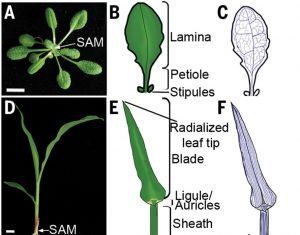 One of the peculiar features of monocots is the presence of sheathing leaf base that encircles the stem, which allow grasses to grow in height, while protecting apical meristem from herbivores by keeping it lower to the ground. Despite its significance, the evolutionary origin has been a controversy for many years. While in 19th century “petiole-sheath” hypothesis was proposed stating grass sheath is equivalent to petiole of eudicot leaves; the 20th century “petiole-leaf” hypothesis suggested that the entire grass leaf is mainly derived from petiole. Using the combination of developmental genetics and computational modelling, Richardson et al. revisited this longstanding unanswered question. The authors built a computational model to understand the grass leaf morphology and to resolve the hypothesis predictions. To gain more insights into the relationship between grass and eudicot leaf, the grass leaf model was modified to produce eudicot leaf, and the assumptions and predictions from the simulations favored the 19th century view of “petiole-sheath” homology. The author highlights that the “key step in grass evolution was the extension of primordium identity and WOX activity along the ad-abaxial boundary to encircle the apex”. (Summary by Prakshi Aneja @PrakshiAneja) Science. 10.1126/science.abf9407
One of the peculiar features of monocots is the presence of sheathing leaf base that encircles the stem, which allow grasses to grow in height, while protecting apical meristem from herbivores by keeping it lower to the ground. Despite its significance, the evolutionary origin has been a controversy for many years. While in 19th century “petiole-sheath” hypothesis was proposed stating grass sheath is equivalent to petiole of eudicot leaves; the 20th century “petiole-leaf” hypothesis suggested that the entire grass leaf is mainly derived from petiole. Using the combination of developmental genetics and computational modelling, Richardson et al. revisited this longstanding unanswered question. The authors built a computational model to understand the grass leaf morphology and to resolve the hypothesis predictions. To gain more insights into the relationship between grass and eudicot leaf, the grass leaf model was modified to produce eudicot leaf, and the assumptions and predictions from the simulations favored the 19th century view of “petiole-sheath” homology. The author highlights that the “key step in grass evolution was the extension of primordium identity and WOX activity along the ad-abaxial boundary to encircle the apex”. (Summary by Prakshi Aneja @PrakshiAneja) Science. 10.1126/science.abf9407
An essential role of MADS-box proteins during wheat spike development
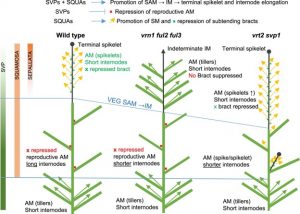 Wheat yields depend on the morphology of the inflorescence known as the spike, which harbors multiple lateral spikelets. To control and manipulate wheat productivity, it is necessary to understand the molecular and genetic mechanisms behind the development of inflorescence architecture. Wheat spike development from the inflorescence meristem (IM) is determined when the terminal spikelet (TS) is formed. The transition from IM to TS largely influences the number of spikelets and eventually the number of grains per spike. Two genes of the SQUAMOSA clade of the MADS-box TF family, i.e., VERNALIZATION1 (VRN1) and FRUITFULL2 (FUL2), are required for this transition, and consequently, spikelets development. In the vrn1ful2 double mutant, the development of TS is abolished, and the axillary meristem generates tiller-like vegetative structures instead of generating spikelets. However, the single mutant (vrn1 or ful2) spikes develop normally with more spikelets due to delayed transition to TS. To explore the regulatory network involved in the spike development, Li et al. (2021) compared the transcriptomes of developing spikes of vrn1 and vrn1ful2 mutants. This led to the identification of three MADS-box genes of SVP clade, i.e., SHORT VEGETATIVE1 (SVP1), SVP2/VEGETATIVE TO REPRODUCTIVE TRANSITION2 (VRT2), and SVP3. Further analysis using genetic interactions among SQUAMOSA and SVP genes revealed their synergistic effects on IM to TS transition but antagonistic effects on the development of spikelets from axillary meristems. SQUAMOSA genes control spikelet development from axillary meristem by repressing SVP genes. Altogether, this SQUAMOSA-SVP cascade is essential for TS development, and the downregulation of SVP genes is required for normal spikelet development. (Summary by Subodh Verma @subodhshanky) Plant Cell 10.1093/plcell/koab243.
Wheat yields depend on the morphology of the inflorescence known as the spike, which harbors multiple lateral spikelets. To control and manipulate wheat productivity, it is necessary to understand the molecular and genetic mechanisms behind the development of inflorescence architecture. Wheat spike development from the inflorescence meristem (IM) is determined when the terminal spikelet (TS) is formed. The transition from IM to TS largely influences the number of spikelets and eventually the number of grains per spike. Two genes of the SQUAMOSA clade of the MADS-box TF family, i.e., VERNALIZATION1 (VRN1) and FRUITFULL2 (FUL2), are required for this transition, and consequently, spikelets development. In the vrn1ful2 double mutant, the development of TS is abolished, and the axillary meristem generates tiller-like vegetative structures instead of generating spikelets. However, the single mutant (vrn1 or ful2) spikes develop normally with more spikelets due to delayed transition to TS. To explore the regulatory network involved in the spike development, Li et al. (2021) compared the transcriptomes of developing spikes of vrn1 and vrn1ful2 mutants. This led to the identification of three MADS-box genes of SVP clade, i.e., SHORT VEGETATIVE1 (SVP1), SVP2/VEGETATIVE TO REPRODUCTIVE TRANSITION2 (VRT2), and SVP3. Further analysis using genetic interactions among SQUAMOSA and SVP genes revealed their synergistic effects on IM to TS transition but antagonistic effects on the development of spikelets from axillary meristems. SQUAMOSA genes control spikelet development from axillary meristem by repressing SVP genes. Altogether, this SQUAMOSA-SVP cascade is essential for TS development, and the downregulation of SVP genes is required for normal spikelet development. (Summary by Subodh Verma @subodhshanky) Plant Cell 10.1093/plcell/koab243.
Functional interaction of nitrate signaling regulator NLP7 and N-degron “recognin” PRT6 enhances abiotic stress tolerance
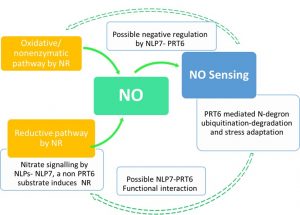 In addition to the vast array of metabolic and signaling roles, nitric oxide (NO) is prominently involved in low oxygen sensing in seeds and waterlogged plants. In Arabidopsis, the cytosolic nitrate reductase (NR) isoforms NIA1/NIA2 catalyze the reductive pathway of NO generation. NO sensing in plants constitute highly regulated proteasomal degradation of ethylene response transcription factors. PROTEOLYSIS6 (PRT6) is a “recognin” that identifies substrates containing N-degron (Cys residue after the initial Met) and polyubiquitinates them. The nitrate-coupled CPK signaling phosphorylates NLP transcription factors (NLP6/ NLP7) and reprograms the expression of downstream N metabolism and signaling genes. To understand the possible crosstalk of N degron proteolysis and nitrate signaling, Castillo et al, 2021, evaluated prt6 nlp7 double mutants and NLP7 over expressing lines in response to different abiotic stresses. Even though NLP7 was not a PRT6 substrate, there were potential cross talks among nitrate signaling, PRT6 and ABA signaling. The “nucleocytoplasmic shuttling” of NLP7, is controlled by NO abundance, posttranslational modifications and possibly by PRT6, offering a regulatory module controlling NO signaling. The possible regulatory role of PRT6 and NLP7 in the synthesis of non-NR derived NO is yet to be addressed. (Summary by Lekshmy Sathee @lekshmysnair) Plant Physiol. https://doi.org/10.1093/plphys/kiab382
In addition to the vast array of metabolic and signaling roles, nitric oxide (NO) is prominently involved in low oxygen sensing in seeds and waterlogged plants. In Arabidopsis, the cytosolic nitrate reductase (NR) isoforms NIA1/NIA2 catalyze the reductive pathway of NO generation. NO sensing in plants constitute highly regulated proteasomal degradation of ethylene response transcription factors. PROTEOLYSIS6 (PRT6) is a “recognin” that identifies substrates containing N-degron (Cys residue after the initial Met) and polyubiquitinates them. The nitrate-coupled CPK signaling phosphorylates NLP transcription factors (NLP6/ NLP7) and reprograms the expression of downstream N metabolism and signaling genes. To understand the possible crosstalk of N degron proteolysis and nitrate signaling, Castillo et al, 2021, evaluated prt6 nlp7 double mutants and NLP7 over expressing lines in response to different abiotic stresses. Even though NLP7 was not a PRT6 substrate, there were potential cross talks among nitrate signaling, PRT6 and ABA signaling. The “nucleocytoplasmic shuttling” of NLP7, is controlled by NO abundance, posttranslational modifications and possibly by PRT6, offering a regulatory module controlling NO signaling. The possible regulatory role of PRT6 and NLP7 in the synthesis of non-NR derived NO is yet to be addressed. (Summary by Lekshmy Sathee @lekshmysnair) Plant Physiol. https://doi.org/10.1093/plphys/kiab382
STOP1-NRT1.1: a new module to optimize nitrogen and growth in acidic media for plants
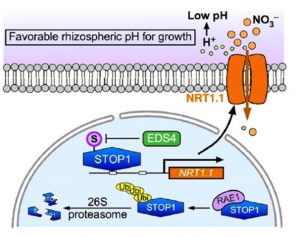 Acidic soils often negatively impact plant growth, yet the application of fertilizers containing urea or ammonia have the effect of acidifying soils. In a recent paper, Ye and colleagues investigated how nitrate uptake through the nitrate transporter NITRATE TRANSPORTER 1.1 (NRT1.1) can mitigate some of the effects of acidic soil. The key players in their study are NRT1.1 and the C2H2-type zinc finger transcription factor SENSITIVE TO PROTON RHIZOTOXICITY1 (STOP1). When the acidity of the soil rises, STOP1 accumulates in the nucleus where it binds to the NRT1.1 promoter and induces higher expression of NRT1.1. Because both nitrate and protons are take up together via NRT1.1, its upregulation leads to a decrease in root-adjacent soil proton levels and thus acidity. The authors are currently trying to understand how plant recycles the protons that are taken up from the cytoplasm, and the answer should lead us to another fascinating tale in understanding plant behavior. (Summary by Sibaji K Sanyal @sibajisanyal ) Plant Cell 10.1093/plcell/koab226
Acidic soils often negatively impact plant growth, yet the application of fertilizers containing urea or ammonia have the effect of acidifying soils. In a recent paper, Ye and colleagues investigated how nitrate uptake through the nitrate transporter NITRATE TRANSPORTER 1.1 (NRT1.1) can mitigate some of the effects of acidic soil. The key players in their study are NRT1.1 and the C2H2-type zinc finger transcription factor SENSITIVE TO PROTON RHIZOTOXICITY1 (STOP1). When the acidity of the soil rises, STOP1 accumulates in the nucleus where it binds to the NRT1.1 promoter and induces higher expression of NRT1.1. Because both nitrate and protons are take up together via NRT1.1, its upregulation leads to a decrease in root-adjacent soil proton levels and thus acidity. The authors are currently trying to understand how plant recycles the protons that are taken up from the cytoplasm, and the answer should lead us to another fascinating tale in understanding plant behavior. (Summary by Sibaji K Sanyal @sibajisanyal ) Plant Cell 10.1093/plcell/koab226
Phosphatidic acid modulates MPK3- and MPK6- mediated hypoxia signaling in Arabidopsis
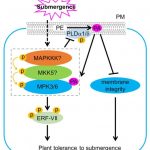
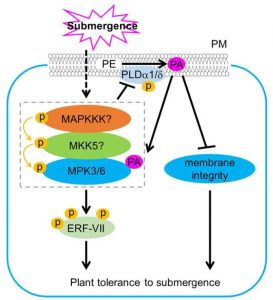 Plants experience changes in gene expression and cellular metabolism to improve their survival to abiotic stresses, such as hypoxia. It has been observed that environmental signals induce phospholipase D (PLD) which generates phosphatidic acid (PA) by cleaving membrane phospholipids. Indeed, submergence induces the accumulation of PA, however, the mechanisms underlying this response have been unknown. In this work, Zhou et al., propose a model based in the PA regulation of submergence tolerance by modulating the membrane integrity and MPK3/6-mediated hypoxia signaling. Submergence generates an accumulation of PA by inducing PLDα1 and PLDδ. PA binds MPK3/6 improving their performance which leads to the phosphorylation of ERFVII, such as RAP2.12, promoting their activities. At the same time, accumulation of PA increases ROS levels which threatens the plant survival. To tackle this harmful situation, MPK3/6 form a regulatory feed-back loop with PLDα1 and PLDδ to reduce their protein levels. In this way, PA is maintained at physiological levels for the plant to face long-term hypoxia conditions. This article sheds light into the mechanisms of hypoxia signaling by reinforcing the role of PA as a key lipid messenger in plant responses to abiotic stresses. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Cell 10.1093/plcell/koab289
Plants experience changes in gene expression and cellular metabolism to improve their survival to abiotic stresses, such as hypoxia. It has been observed that environmental signals induce phospholipase D (PLD) which generates phosphatidic acid (PA) by cleaving membrane phospholipids. Indeed, submergence induces the accumulation of PA, however, the mechanisms underlying this response have been unknown. In this work, Zhou et al., propose a model based in the PA regulation of submergence tolerance by modulating the membrane integrity and MPK3/6-mediated hypoxia signaling. Submergence generates an accumulation of PA by inducing PLDα1 and PLDδ. PA binds MPK3/6 improving their performance which leads to the phosphorylation of ERFVII, such as RAP2.12, promoting their activities. At the same time, accumulation of PA increases ROS levels which threatens the plant survival. To tackle this harmful situation, MPK3/6 form a regulatory feed-back loop with PLDα1 and PLDδ to reduce their protein levels. In this way, PA is maintained at physiological levels for the plant to face long-term hypoxia conditions. This article sheds light into the mechanisms of hypoxia signaling by reinforcing the role of PA as a key lipid messenger in plant responses to abiotic stresses. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Cell 10.1093/plcell/koab289