Plant Science Research Weekly: January 21, 2022
TT2 controls rice thermotolerance through SCT1-dependent alteration of wax biosynthesis
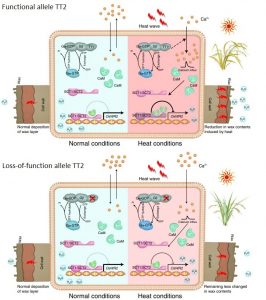 Global warming severely affects agricultural harvests, risking food security. To deal with heat stress, plants show different strategies. Indeed, heat increases intracellular Ca2+ levels to activate a heat shock response. In addition, GTP-binding proteins, which transduce extracellular signals to intracellular effectors, regulate Ca2+ currents when plants face abiotic stresses through calmodulin-binding proteins (CaM). In this work, Kan et al., identified a quantitative trait locus (QTL) TT2 (THERMOTOLERANCE 2) which encodes for a G-protein subunit (Gγ) and that provides thermotolerance in rice. When rice plants carry the natural allele of disrupted TT2 they show less reductions in wax content compared with the plants carrying the non-disrupted allele of TT2. Furthermore, plants with disrupted the TT2 allele show an attenuated Ca2+ spike which reduces the Ca2+-dependent interaction beteeen CaM and the transcription factor SCT1 (Sensing Ca2+ Transcription factor 1). A downstream target of SCT1 is Wax Synthesis Regulatory 2 (OsWR2), thus leading to the reduction of wax content. This work increases the number of available genetic tools to develop tolerant crops to heat stress. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Nature Plants. 10.1038/s41477-021-01039-0
Global warming severely affects agricultural harvests, risking food security. To deal with heat stress, plants show different strategies. Indeed, heat increases intracellular Ca2+ levels to activate a heat shock response. In addition, GTP-binding proteins, which transduce extracellular signals to intracellular effectors, regulate Ca2+ currents when plants face abiotic stresses through calmodulin-binding proteins (CaM). In this work, Kan et al., identified a quantitative trait locus (QTL) TT2 (THERMOTOLERANCE 2) which encodes for a G-protein subunit (Gγ) and that provides thermotolerance in rice. When rice plants carry the natural allele of disrupted TT2 they show less reductions in wax content compared with the plants carrying the non-disrupted allele of TT2. Furthermore, plants with disrupted the TT2 allele show an attenuated Ca2+ spike which reduces the Ca2+-dependent interaction beteeen CaM and the transcription factor SCT1 (Sensing Ca2+ Transcription factor 1). A downstream target of SCT1 is Wax Synthesis Regulatory 2 (OsWR2), thus leading to the reduction of wax content. This work increases the number of available genetic tools to develop tolerant crops to heat stress. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Nature Plants. 10.1038/s41477-021-01039-0
After all, mutations are not that random
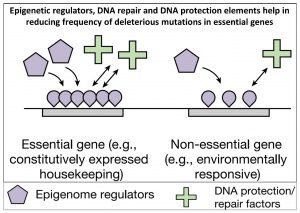 Mutations, defined as changes in DNA sequence, have long been considered to be random. However, growing evidence suggests that maybe mutations are not random, but instead some loci in the genome are hot spots for mutations, while other loci rarely mutate. This effect is considered mutation bias. A recent study by Monroe et al. in Arabidopsis thaliana supports this model. The authors observed that mutations are more common in intergenic regions than in the genes. Similarly, they also observed a high probability of mutations in regions associated with environmental adaptation and a low probability of mutation in genes associated with core biological functions. This is consistent with the notion that genes involved in environmental adaptation/responses are evolving and diversifying faster than genes involved in core biological processes. The authors also observed this mutation bias is associated with other factors such as DNA repair and histone modifications, which seems to favor a low frequency of mutation in essential genes as compared to other genes. The authors draw a comparison between these effects and loaded dice, which act to favor a particular outcome (beneficial mutations) and are biased against other outcomes (harmful mutations). Summary by Kamal Kumar Malukani @KamalMalukani. Nature 10.1038/s41586-021-04269-6
Mutations, defined as changes in DNA sequence, have long been considered to be random. However, growing evidence suggests that maybe mutations are not random, but instead some loci in the genome are hot spots for mutations, while other loci rarely mutate. This effect is considered mutation bias. A recent study by Monroe et al. in Arabidopsis thaliana supports this model. The authors observed that mutations are more common in intergenic regions than in the genes. Similarly, they also observed a high probability of mutations in regions associated with environmental adaptation and a low probability of mutation in genes associated with core biological functions. This is consistent with the notion that genes involved in environmental adaptation/responses are evolving and diversifying faster than genes involved in core biological processes. The authors also observed this mutation bias is associated with other factors such as DNA repair and histone modifications, which seems to favor a low frequency of mutation in essential genes as compared to other genes. The authors draw a comparison between these effects and loaded dice, which act to favor a particular outcome (beneficial mutations) and are biased against other outcomes (harmful mutations). Summary by Kamal Kumar Malukani @KamalMalukani. Nature 10.1038/s41586-021-04269-6
The genomic ecosystem of transposable elements in maize
 A new paper looking at transposable elements in maize uses the framework that genomes are similar to ecosystems, in that it is essential to study them comprehensively, from the level of each element to the global structure. Transposable elements (TEs) are dynamic and persistent within plant genomes. Understanding their behavior and physiology can give us a broader perspective of the evolutionary potential of their host genome and species. Stitzer and colleagues explored the Maize B73 reference genome annotation to characterize TE families and their relationships. They annotated and classified more than 27,000 TE families in 13 superfamilies. To build a family survival (age) model, they measured features like TE taxonomy, length, expression, methylation, etc. Many characteristics were shared across families of the same superfamily. Nevertheless, some families had specific contributions to the community. Further, some features correlated to TE age and led the model response. Predictions emphasize studying each family apart since they are adapted to a unique niche. TEs evolve in parallel to maize and are vital components of its evolution and physiology. This work encourages integrated TE ecosystem depiction in our current plant genome analysis. (Summary and sketch by Eddy Mendoza-Galindo @IamSomnya) PLOS Genetics 10.1371/journal.pgen.100976
A new paper looking at transposable elements in maize uses the framework that genomes are similar to ecosystems, in that it is essential to study them comprehensively, from the level of each element to the global structure. Transposable elements (TEs) are dynamic and persistent within plant genomes. Understanding their behavior and physiology can give us a broader perspective of the evolutionary potential of their host genome and species. Stitzer and colleagues explored the Maize B73 reference genome annotation to characterize TE families and their relationships. They annotated and classified more than 27,000 TE families in 13 superfamilies. To build a family survival (age) model, they measured features like TE taxonomy, length, expression, methylation, etc. Many characteristics were shared across families of the same superfamily. Nevertheless, some families had specific contributions to the community. Further, some features correlated to TE age and led the model response. Predictions emphasize studying each family apart since they are adapted to a unique niche. TEs evolve in parallel to maize and are vital components of its evolution and physiology. This work encourages integrated TE ecosystem depiction in our current plant genome analysis. (Summary and sketch by Eddy Mendoza-Galindo @IamSomnya) PLOS Genetics 10.1371/journal.pgen.100976
Transcriptome sequencing of a novel albino mutant of hexaploid sweetpotato
 Albino plants lack chlorophyll, which means that they cannot photosynthesize. These kinds of plant are unique and can offer information as to the molecular mechanism of chlorophyll degradation and photosynthesis in plants. Here, Arisha et al. explored the differences in gene expression patterns of seedling leaf tissues from albino mutant sweetpotatoes (Ipomoea batatas) and their green counterparts, using RNA sequencing. In albino plants compared to the green plants, the authors discovered that 3000 unigenes involved in plant mobilization of stored carbohydrates and fats were highly expressed while 2418 unigenes involved in chloroplast development, biosynthesis and differentiation were downregulated. Similarly, genes involved in the metabolism of chlorophyll a, b and carotenoid pigments were downregulated in albino plants compared to the green plants. Through this study, the authors successfully interrogated the complex hexaploid sweetpotato genome and identified important gene regulatory networks. This work also provided insight into some complex biological process for photosynthesis in plants. (Summary by Modesta Abugu @modestannedi) Plant Mol. Biol. Rep. 10.1007/s11105-020-01239-6
Albino plants lack chlorophyll, which means that they cannot photosynthesize. These kinds of plant are unique and can offer information as to the molecular mechanism of chlorophyll degradation and photosynthesis in plants. Here, Arisha et al. explored the differences in gene expression patterns of seedling leaf tissues from albino mutant sweetpotatoes (Ipomoea batatas) and their green counterparts, using RNA sequencing. In albino plants compared to the green plants, the authors discovered that 3000 unigenes involved in plant mobilization of stored carbohydrates and fats were highly expressed while 2418 unigenes involved in chloroplast development, biosynthesis and differentiation were downregulated. Similarly, genes involved in the metabolism of chlorophyll a, b and carotenoid pigments were downregulated in albino plants compared to the green plants. Through this study, the authors successfully interrogated the complex hexaploid sweetpotato genome and identified important gene regulatory networks. This work also provided insight into some complex biological process for photosynthesis in plants. (Summary by Modesta Abugu @modestannedi) Plant Mol. Biol. Rep. 10.1007/s11105-020-01239-6
The PARTHENOGENESIS gene that contributes the clonal propagation through seeds in dicot dandelion
 In plants with sexual reproduction, the male contribution (two sperm cells) is necessary to fertilize two female cells, the egg cell that gives rise to the embryo and the central cell that gives rise to the endosperm. In contrast, apomictic plants (0.1% of flowering plants spread over 120 genera) don´t need the male input, they generate seeds asexually with only maternal genetic information. This trait ensures that the progeny is genetically identical to the maternal plant, and ensures seed production even without pollination, so is particularly interesting for seed crop production. Dandelion (Taraxacum officinale) usually reproduce asexually through apomixis, specifically parthenogenesis in which the unfertilized egg cell undergoes embryogenesis. Previously in dadelion the PARTHENOGENESIS (PAR) locus was identified that has a dominant effect on apomixis. Here, Underwood, Vijverberg, Rigola, et al. assembled and analysed the sequences of the PAR locus of both triploid apomictic and diploid sexual forms of Taraxacum. They noticed that the PAR gene is expressed in the egg cells of the apomictic dandelion whereas in the sexual form the PAR gene is mainly expressed in pollen. Further, the authors found a 1kb insertion upstream the PAR promoter region in the apomictic plants but absent in the sexual ones. Finally, they confirmed the functional importance of the PAR allele; driving the expression of a homologous lettuce gene (Lssex) in ovules was sufficient to induce parthenogenesis. These findings will contribute to the implementation of synthetic apomixis in dicot plants. (Summary by Rigel Salinas-Gamboa @Rigelitactica). Nature Genetics 10.1038/s41588-021-00984-y
In plants with sexual reproduction, the male contribution (two sperm cells) is necessary to fertilize two female cells, the egg cell that gives rise to the embryo and the central cell that gives rise to the endosperm. In contrast, apomictic plants (0.1% of flowering plants spread over 120 genera) don´t need the male input, they generate seeds asexually with only maternal genetic information. This trait ensures that the progeny is genetically identical to the maternal plant, and ensures seed production even without pollination, so is particularly interesting for seed crop production. Dandelion (Taraxacum officinale) usually reproduce asexually through apomixis, specifically parthenogenesis in which the unfertilized egg cell undergoes embryogenesis. Previously in dadelion the PARTHENOGENESIS (PAR) locus was identified that has a dominant effect on apomixis. Here, Underwood, Vijverberg, Rigola, et al. assembled and analysed the sequences of the PAR locus of both triploid apomictic and diploid sexual forms of Taraxacum. They noticed that the PAR gene is expressed in the egg cells of the apomictic dandelion whereas in the sexual form the PAR gene is mainly expressed in pollen. Further, the authors found a 1kb insertion upstream the PAR promoter region in the apomictic plants but absent in the sexual ones. Finally, they confirmed the functional importance of the PAR allele; driving the expression of a homologous lettuce gene (Lssex) in ovules was sufficient to induce parthenogenesis. These findings will contribute to the implementation of synthetic apomixis in dicot plants. (Summary by Rigel Salinas-Gamboa @Rigelitactica). Nature Genetics 10.1038/s41588-021-00984-y
Fast and global reorganization of the chloroplast protein biogenesis network during heat acclimation
 With the rising climatological extremes, heat stress is a major concern towards sustainable crop yield and productivity as it impairs several physiological and developmental processes. Due to the sessile lifestyle of land plants, they undergo various acclimation responses to cope with fluctuating temperatures. Chloroplasts, apart from being the energy-producing organelles, are also a central player for numerous acclimation responses; hence they are also regarded as the “hub of acclimation”. However, there is very limited understanding about the regulatory mechanisms responsible for heat acclimation of chloroplasts. Trösch et al. revealed an altered pattern of plastid protein biosynthesis and translation elongation as the major heat acclimation processes through time-resolved transcript and ribosome profiling. The authors highlight that these acclimation responses are conserved between Chlamydomonas and tobacco. The authors have also identified certain ribosome-associated factors suggesting that the co-translational action of these factors might be crucial for heat acclimation. Further investigation will be required to elucidate the detailed mechanism underlying co-translational processes during heat acclimation. (Summary by Prakshi Aneja @PrakshiAneja) Plant Cell 10.1093/plcell/koab317
With the rising climatological extremes, heat stress is a major concern towards sustainable crop yield and productivity as it impairs several physiological and developmental processes. Due to the sessile lifestyle of land plants, they undergo various acclimation responses to cope with fluctuating temperatures. Chloroplasts, apart from being the energy-producing organelles, are also a central player for numerous acclimation responses; hence they are also regarded as the “hub of acclimation”. However, there is very limited understanding about the regulatory mechanisms responsible for heat acclimation of chloroplasts. Trösch et al. revealed an altered pattern of plastid protein biosynthesis and translation elongation as the major heat acclimation processes through time-resolved transcript and ribosome profiling. The authors highlight that these acclimation responses are conserved between Chlamydomonas and tobacco. The authors have also identified certain ribosome-associated factors suggesting that the co-translational action of these factors might be crucial for heat acclimation. Further investigation will be required to elucidate the detailed mechanism underlying co-translational processes during heat acclimation. (Summary by Prakshi Aneja @PrakshiAneja) Plant Cell 10.1093/plcell/koab317
The splicing factor RNA BINDING PROTEIN 45d regulates temperature-responsive flowering
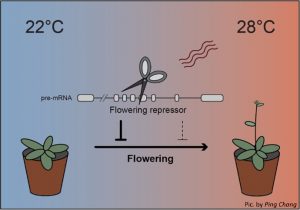 Posttranscriptional events such as splicing are part of the gene regulation toolbox. Previous studies have demonstrated a role for U-rich small nuclear ribonucleoproteins (snRNPs) such as U1 in the control of splicing. In a recent paper in The Plant Cell, Chang et al. identified a splicing factor, RNA BINDING PROTEIN 45d (RBP45d), in Arabidopsis, that is involved in pre-mRNA splicing. RBP45d together with another U1 snRNP component PRP39a, regulate alternative splicing of FLOWERING LOCUS M (FLM), which is a key flowering repressor under low temperature. Rising ambient temperature promotes flowering through the alternative splicing of FLM, which leads to the degradation of this flowering repressor. In summary, the authors identified and characterized an U1 snRNP component RNA BINDING PROTEIN 45d, which regulates temperature-responsive flowering in Arabidopsis. (Summary by Chandan Kumar Gautam @chandan_gautam) Plant Cell 10.1093/plcell/koab273
Posttranscriptional events such as splicing are part of the gene regulation toolbox. Previous studies have demonstrated a role for U-rich small nuclear ribonucleoproteins (snRNPs) such as U1 in the control of splicing. In a recent paper in The Plant Cell, Chang et al. identified a splicing factor, RNA BINDING PROTEIN 45d (RBP45d), in Arabidopsis, that is involved in pre-mRNA splicing. RBP45d together with another U1 snRNP component PRP39a, regulate alternative splicing of FLOWERING LOCUS M (FLM), which is a key flowering repressor under low temperature. Rising ambient temperature promotes flowering through the alternative splicing of FLM, which leads to the degradation of this flowering repressor. In summary, the authors identified and characterized an U1 snRNP component RNA BINDING PROTEIN 45d, which regulates temperature-responsive flowering in Arabidopsis. (Summary by Chandan Kumar Gautam @chandan_gautam) Plant Cell 10.1093/plcell/koab273
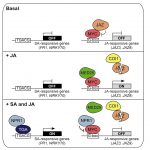
Suppression of MYC transcription activators by the immune cofactor NPR1 fine-tunes plant immune responses
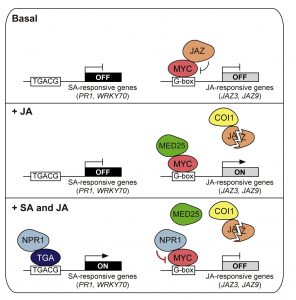 In Arabidopsis, NONEXPRESSOR OF PR GENES 1 (NPR1) plays an important role in the antagonistic crosstalk of salicylic acid (SA) and jasmonic acid (JA) signalling. It activates SA-induced genes that protect against biotrophic pathogens and suppresses JA-induced genes that protect against necrotrophic pathogens. The biotrophic Pseudomonas syringae pv. tomato overcomes the plant’s SA-induced immune response by producing the toxin coronatine (COR), which mimics biologically active JA that suppresses SA-induced immunity. Here, Nomoto et al. show the molecular mechanism whereby SA-induced NPR1 in turn negatively regulates JA-responsive genes. Through in vitro and in vivo experiments, the authors demonstrate that NPR1 interacts with the transcription factor MYC2, preventing it from associating with the MED25 subunit of the Mediator transcriptional co-activator complex and performing its role as a transcriptional activator of JA-induced genes. RNAseq and ChIP-seq experiments reveal that NPR1 targets JA-responsive promoters, co-occupied by transcriptionally active MYC2-MED25 complexes. ChIP experiments also show how SA-signalling does not prevent MYC2 binding to JA-responsive promoters but does prevent MED25 accumulation. Using Pseudomonas syringae pathogen assays, the authors demonstrate how NPR1 suppresses MYC activators to overcome COR-mediated infection. (Summary by Jiawen Chen @Jiaaawen) Cell Reports 10.1016/j.celrep.2021.110125
In Arabidopsis, NONEXPRESSOR OF PR GENES 1 (NPR1) plays an important role in the antagonistic crosstalk of salicylic acid (SA) and jasmonic acid (JA) signalling. It activates SA-induced genes that protect against biotrophic pathogens and suppresses JA-induced genes that protect against necrotrophic pathogens. The biotrophic Pseudomonas syringae pv. tomato overcomes the plant’s SA-induced immune response by producing the toxin coronatine (COR), which mimics biologically active JA that suppresses SA-induced immunity. Here, Nomoto et al. show the molecular mechanism whereby SA-induced NPR1 in turn negatively regulates JA-responsive genes. Through in vitro and in vivo experiments, the authors demonstrate that NPR1 interacts with the transcription factor MYC2, preventing it from associating with the MED25 subunit of the Mediator transcriptional co-activator complex and performing its role as a transcriptional activator of JA-induced genes. RNAseq and ChIP-seq experiments reveal that NPR1 targets JA-responsive promoters, co-occupied by transcriptionally active MYC2-MED25 complexes. ChIP experiments also show how SA-signalling does not prevent MYC2 binding to JA-responsive promoters but does prevent MED25 accumulation. Using Pseudomonas syringae pathogen assays, the authors demonstrate how NPR1 suppresses MYC activators to overcome COR-mediated infection. (Summary by Jiawen Chen @Jiaaawen) Cell Reports 10.1016/j.celrep.2021.110125