GWAS and the fight against darkness-induced starvation
By Feng Zhu and Alisdair R. Fernie
Background: In cellular circumstances under which nutrients are scarce, plants must degrade polymers to recycle carbon (C) and nitrogen (N) sources for use in their sink tissues. Dark-induced senescence is one such circumstance. During dark-induced senescence, metabolism is dramatically reprogrammed to lengthen the plant lifespan by switching from anabolic to catabolic processes and recycling and remobilizing C and N sources. Previous research largely focused on individual genes or pathways by characterizing their mutants or one stage of this process, but our understanding of metabolite levels are regulated during this process remains fragmentary.
Question: We wanted to know the global genomic landscape of the metabolic shifts underpinning dark-induced senescence. We thus adopted a time-resolved genome-wide association-based approach to characterize the variation in photochemical efficiency and contents of primary and lipid metabolites at the beginning, after 3 and 6 days in darkness.
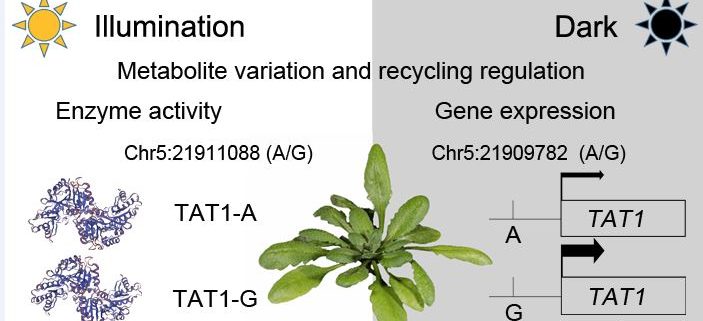
Findings: The metabolites directly involved in energy generation (sugars, sugar alcohols and the organic acids of the TCA cycle) decreased first and may act as early-responding metabolites for recycling. Alternative respiration substrates (such as tyrosine, threonine and BCAAs) whose contents first increased and then decreased, may be late-responding metabolites to supply energy under extended darkness. Moreover, we identified 215 associations with 81 candidate genes. Different lead SNPs for the association between tyrosine and TYROSINE AMINOTRANSFERASE 1 (TAT1) affected its enzyme activity and gene expression under normal and dark conditions, respectively. SNP variation affecting the expression of THREONINE ALDOLASE 1 (THA1) and the amino acid transporter gene AVT1B explained the variation in threonine and glycine levels, respectively, in the dark.
Next steps: Although dark-induced senescence is complex, we demonstrated the great power and high accuracy of time-resolved mGWAS by the identification of many associations and the detailed genetic effects on candidate genes in the present study. With the development of high-throughput sequencing methods, other genetic variation information (such as large fragment deletions and insertions, chromosomal rearrangements) are available; their combination with time-resolved mGWAS will likely provide more comprehensive understanding of the plant genetic and metabolic landscapes and promote the dissection of complex phenotypes.
Reference:
Feng Zhu, Saleh Alseekh, Kaan Koper, Hao Tong, Zoran Nikoloski, Thomas Naake, Haijun Liu, Jianbing Yan, Yariv Brotman, Weiwei Wen, Hiroshi Maeda, Yunjiang Cheng*, Alisdair R. Fernie* (2021). Genome-wide association of the metabolic shifts underpinning dark-induced senescence in Arabidopsis. https://doi.org/10.1093/plcell/koab251