Plant Science Research Weekly: January 10
Review. Diatom molecular research comes of age: Model species for studying phytoplankton biology and diversity
 Diatoms are photosynthetic eukaryotes and contribute substantially to global carbon fixation. They are distantly related to green plants, having shared the same primary endosymbiotic event, although they subsequently underwent additional secondary endosymbioses. There are over 100,000 species of diatoms, of which two have been the greatest focus for genomic efforts. Falciatore et al. review current and future diatom research objectives. The article reviews the evolution and phylogeny of diatoms, their cell division and life cycle, and insights from genomic studies. The authors also review the processes of photosynthesis and nutrient uptake in diatoms and their environmental responses, and discuss them in comparison to the green algae and plants. (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00158
Diatoms are photosynthetic eukaryotes and contribute substantially to global carbon fixation. They are distantly related to green plants, having shared the same primary endosymbiotic event, although they subsequently underwent additional secondary endosymbioses. There are over 100,000 species of diatoms, of which two have been the greatest focus for genomic efforts. Falciatore et al. review current and future diatom research objectives. The article reviews the evolution and phylogeny of diatoms, their cell division and life cycle, and insights from genomic studies. The authors also review the processes of photosynthesis and nutrient uptake in diatoms and their environmental responses, and discuss them in comparison to the green algae and plants. (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00158
Review: Role of blue and red light in stomatal dynamic behaviour
 Guard cells are extremely sensitive and dynamic, and their behaviour controls rates of gas exchange and transpiration, which affect evaporative cooling and transport in the xylem. Matthews et al. review the roles of light signalling pathways in guard cell responses. Cues that control guard cell ion channels and transporters (hence stomatal pore size) include temperature, CO2 and vapor-pressure deficit, as well as both blue and red light. The blue-light response is mediated by phototropins, whereas the red-light response is a consequence of photosynthesis in the guard cells and mesophyll cells, mediated by changes in the concentrations of CO2 , ATP, sugars, and other metabolites. In some situations, the blue light response increases transpiration without increasing photosynthesis, so tuning guard cell responses to different wavelengths of light might be a way to improve water-use efficiency or cooling, as described in the article. (Summary by Mary Williams) J. Exp. Bot. 10.1093/jxb/erz563
Guard cells are extremely sensitive and dynamic, and their behaviour controls rates of gas exchange and transpiration, which affect evaporative cooling and transport in the xylem. Matthews et al. review the roles of light signalling pathways in guard cell responses. Cues that control guard cell ion channels and transporters (hence stomatal pore size) include temperature, CO2 and vapor-pressure deficit, as well as both blue and red light. The blue-light response is mediated by phototropins, whereas the red-light response is a consequence of photosynthesis in the guard cells and mesophyll cells, mediated by changes in the concentrations of CO2 , ATP, sugars, and other metabolites. In some situations, the blue light response increases transpiration without increasing photosynthesis, so tuning guard cell responses to different wavelengths of light might be a way to improve water-use efficiency or cooling, as described in the article. (Summary by Mary Williams) J. Exp. Bot. 10.1093/jxb/erz563
Combinatorial interactions of the LEC1 transcription factor specify diverse developmental programs during soybean seed development ($)
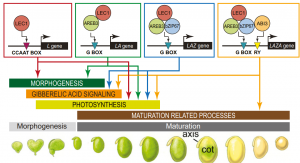 LEC1 (LEAFY COTYLEDON 1) transcriptional factor regulates multiple developmental processes during different stages of seed development including macromolecule accumulation, inhibition of germination and morphogenesis. In this paper Jo et al., have uncovered the mechanism of LEC1 functioning with other transcriptional factors through co-expression analysis of genes to regulate downstream gene expression to fine-tune seed development. LEC1 in combination with ABA-RESPONSIVE ELEMENT BINDING PROTEIN3 (AREB3) is shown to regulate morphogenesis. Along with multiple partners like BASIC LEUCINE ZIPPER67 (bZIP67), AREB3 and ABA INSENSITIVE3 (ABI3), LEC1 regulates the photosynthesis which is initiated post-seed morphogenesis. Finally, the combination of all these transcriptional factors was identified to contribute towards the seed maturation. The authors also identified cis-regulatory modules for the transcriptional factors and validated by expressing the cis-region to minimal 35s Promoter tagged to LUCIFERASE in cotyledon protoplasts from EM-stage embryos. Overall this paper has shown the mechanism of an atypical NF-YB transcriptional factor, LEC1 regulating a multitude of seed developmental processes in soybean. (Summary by Suresh Damodaran) 10.1073/pnas.1918441117
LEC1 (LEAFY COTYLEDON 1) transcriptional factor regulates multiple developmental processes during different stages of seed development including macromolecule accumulation, inhibition of germination and morphogenesis. In this paper Jo et al., have uncovered the mechanism of LEC1 functioning with other transcriptional factors through co-expression analysis of genes to regulate downstream gene expression to fine-tune seed development. LEC1 in combination with ABA-RESPONSIVE ELEMENT BINDING PROTEIN3 (AREB3) is shown to regulate morphogenesis. Along with multiple partners like BASIC LEUCINE ZIPPER67 (bZIP67), AREB3 and ABA INSENSITIVE3 (ABI3), LEC1 regulates the photosynthesis which is initiated post-seed morphogenesis. Finally, the combination of all these transcriptional factors was identified to contribute towards the seed maturation. The authors also identified cis-regulatory modules for the transcriptional factors and validated by expressing the cis-region to minimal 35s Promoter tagged to LUCIFERASE in cotyledon protoplasts from EM-stage embryos. Overall this paper has shown the mechanism of an atypical NF-YB transcriptional factor, LEC1 regulating a multitude of seed developmental processes in soybean. (Summary by Suresh Damodaran) 10.1073/pnas.1918441117
The Arabidopsis receptor kinase IRK is polarized and represses specific cell division ($)
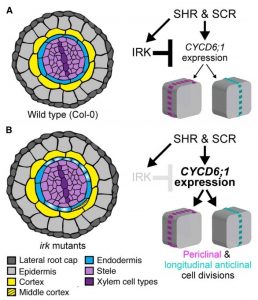 Orientation of cell division decides daughter cell fate and is fundamentally important for tissue patterning and morphology. For instance, asymmetric cell division leads to the generation of new cell types; in contrast, symmetric division produces cells with similar identity in a proliferative manner. Campos et al. studied proteins involved in tissue patterning in Arabidopsis thaliana root. They identified INFLORESCENCE AND ROOT APICES RECEPTOR KINASE (IRK), a LRR-RLKs that is polarized in the ground tissue and important for its patterning. IRK localizes into distinct plasma membrane domains in various cell types depending on the radially adjacent cells, and irk mutants show radially abnormal cell division in the endodermis. In the signaling cascade, IRK downregulates CYCD6;1 and regulates endodermal cell division. As a result, in the irk mutant, the repression of CYCD6;1 through IRK is inactive and causing abnormal endodermal cell division in the radial axis. Altogether, this study provided the mechanistic explanation on ground tissue patterning through polarly localized IRK protein. (Summary by Arif Ashraf) Developmental Cell 10.1016/j.devcel.2019.12.001
Orientation of cell division decides daughter cell fate and is fundamentally important for tissue patterning and morphology. For instance, asymmetric cell division leads to the generation of new cell types; in contrast, symmetric division produces cells with similar identity in a proliferative manner. Campos et al. studied proteins involved in tissue patterning in Arabidopsis thaliana root. They identified INFLORESCENCE AND ROOT APICES RECEPTOR KINASE (IRK), a LRR-RLKs that is polarized in the ground tissue and important for its patterning. IRK localizes into distinct plasma membrane domains in various cell types depending on the radially adjacent cells, and irk mutants show radially abnormal cell division in the endodermis. In the signaling cascade, IRK downregulates CYCD6;1 and regulates endodermal cell division. As a result, in the irk mutant, the repression of CYCD6;1 through IRK is inactive and causing abnormal endodermal cell division in the radial axis. Altogether, this study provided the mechanistic explanation on ground tissue patterning through polarly localized IRK protein. (Summary by Arif Ashraf) Developmental Cell 10.1016/j.devcel.2019.12.001
Promotion of BR biosynthesis by miR444 is required for ammonium-triggered inhibition of root growth ($)
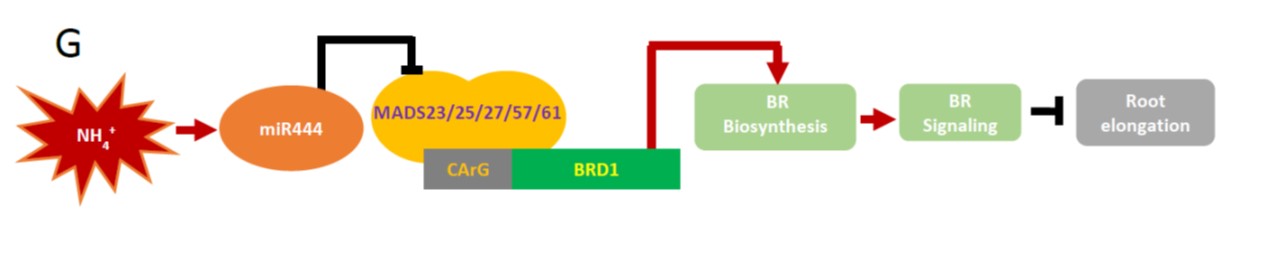 Nitrogen, as one of the most important plant nutrients, profoundly affects root growth. Nitrogen is usually taken up in one of two forms, nitrate (NO3–) or ammonium (NH4+), and the two forms affect root growth differently. Here, Jiao et al. demonstrate a connection between ammonium, a small RNA, and brassinosteroid hormones. Specifically, they show that NH4+ induces miR444, which targets MADS-box transcription repressors that repress expression of a BR synthesis gene, leading to increased levels of brassinosteroids, and root growth inhibition. Similarly, adding BRZ, an inhibitor of brassinosteroid synthesis, to the growth medium alleviated the repression induced by NH4+, showing that ammonium-inhibition of root growth is mediated through brassinosteroids. Given that ammonium is a preferred nitrogen source for rice, this finding may lead to strategies to enhance root growth. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.00190
Nitrogen, as one of the most important plant nutrients, profoundly affects root growth. Nitrogen is usually taken up in one of two forms, nitrate (NO3–) or ammonium (NH4+), and the two forms affect root growth differently. Here, Jiao et al. demonstrate a connection between ammonium, a small RNA, and brassinosteroid hormones. Specifically, they show that NH4+ induces miR444, which targets MADS-box transcription repressors that repress expression of a BR synthesis gene, leading to increased levels of brassinosteroids, and root growth inhibition. Similarly, adding BRZ, an inhibitor of brassinosteroid synthesis, to the growth medium alleviated the repression induced by NH4+, showing that ammonium-inhibition of root growth is mediated through brassinosteroids. Given that ammonium is a preferred nitrogen source for rice, this finding may lead to strategies to enhance root growth. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.00190
Pathogenic bacteria target plant plasmodesmata to colonize and invade surrounding tissues
 Plasmodesmata are regulated channels that connect adjacent cells, allowing movement of metabolites, RNA, proteins, and pathogens. Plants close their plasmodesmata as part of their immune response, but this closure can be interfered with by pathogens. Aung et al. examined the repertoire of effector proteins produced by the plant pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 and found that one of these, HopO1-1, localizes to the plasmodesmata. When HopO1-1 is deleted or compromised, the bacteria are severely compromised in virulence, and movement of fluorescent proteins through the plasmodemata is also diminished as compared to in the presence of the wild-type HopO1-1. PD-located receptor-like proteins (PDLPs) are known to regulate callose deposition at the plasmodesmata, which affects their permeability. The authors found that HopO1-1 interacts with two members of this family, PDLP 5 and PDLP7 and affects their stability, potentially through ribosylation. Knocking out these genes makes plants more susceptible to bacteria, consistent with a model in which the bacterial effector HopO1-1 enhances its virulence through targeting these proteins. (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00707
Plasmodesmata are regulated channels that connect adjacent cells, allowing movement of metabolites, RNA, proteins, and pathogens. Plants close their plasmodesmata as part of their immune response, but this closure can be interfered with by pathogens. Aung et al. examined the repertoire of effector proteins produced by the plant pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 and found that one of these, HopO1-1, localizes to the plasmodesmata. When HopO1-1 is deleted or compromised, the bacteria are severely compromised in virulence, and movement of fluorescent proteins through the plasmodemata is also diminished as compared to in the presence of the wild-type HopO1-1. PD-located receptor-like proteins (PDLPs) are known to regulate callose deposition at the plasmodesmata, which affects their permeability. The authors found that HopO1-1 interacts with two members of this family, PDLP 5 and PDLP7 and affects their stability, potentially through ribosylation. Knocking out these genes makes plants more susceptible to bacteria, consistent with a model in which the bacterial effector HopO1-1 enhances its virulence through targeting these proteins. (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00707
Resistant tomato restricts colonization and invasion by the pathogen Ralstonia solanacearum at four organismal levels
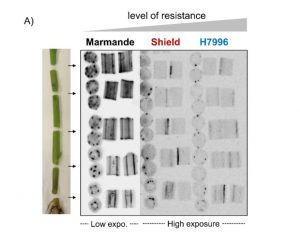 Ralstonia solanacearum is a pathogenic bacterium that infects many important crop species, including tomato. Following invasion into the roots, the bacteria move upwards into the shoot and cause dramatic wilting. Previous studies have identified moderately and highly resistant lines. Here, Planas-Marquès et al. used luminescent bacteria and a grafting method to investigate how the genetic resistance controls pathogen entry and spread. They found that multiple locations are crucial for these processes, including the junction of root and shoot, verticle movement within the xylem, circular spread between vessels and radial spread from the vessels into shoot tissues. Successful pathogens must surmount each of these barriers. The authors discuss how resistant plants block this progression, for example through obstruction of xylem vessels by gums and tyloses. (Summary by Mary Williams) J. Exp. Bot. 10.1093/jxb/erz562
Ralstonia solanacearum is a pathogenic bacterium that infects many important crop species, including tomato. Following invasion into the roots, the bacteria move upwards into the shoot and cause dramatic wilting. Previous studies have identified moderately and highly resistant lines. Here, Planas-Marquès et al. used luminescent bacteria and a grafting method to investigate how the genetic resistance controls pathogen entry and spread. They found that multiple locations are crucial for these processes, including the junction of root and shoot, verticle movement within the xylem, circular spread between vessels and radial spread from the vessels into shoot tissues. Successful pathogens must surmount each of these barriers. The authors discuss how resistant plants block this progression, for example through obstruction of xylem vessels by gums and tyloses. (Summary by Mary Williams) J. Exp. Bot. 10.1093/jxb/erz562
Medicago-Sinorhizobium-Ralstonia co-infection reveals legume nodules as pathogen confined infection sites developing weak defenses
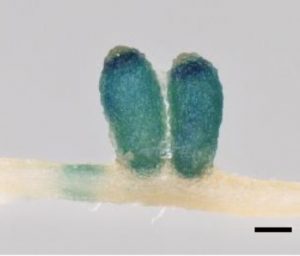 The pathogenic bacterium Ralstonia solanacearum enters roots through wounds and also at root tips. It can also infect legume nodules. Benezech et al. investigated how this infection occurs, and how it is affected by and affects nitrogen fixation. The authors found that nodules are as permissive of Ralstonia infection as root tips, and that this infection is not diminished by nitrogen fixation. Optimal pathogen growth in the nodules depends on the presence of the type-III secretion system, the structure though which bacteria introduce effector proteins into their hosts. This finding suggest that the nodule tissues mount a defense against the pathogen, which was supported by transcriptomic data. These data also show that the defense response in nodules is weaker and distinct from that of root tissue. Interestingly, the increased susceptibility of nodules to infection is counter-balanced by an unknown system that prevents dissemination of bacteria, both symbiotic and pathogenic, from the nodules and into the root system. (Summary by Mary Williams) Curr. Biol. 10.1016/j.cub.2019.11.066
The pathogenic bacterium Ralstonia solanacearum enters roots through wounds and also at root tips. It can also infect legume nodules. Benezech et al. investigated how this infection occurs, and how it is affected by and affects nitrogen fixation. The authors found that nodules are as permissive of Ralstonia infection as root tips, and that this infection is not diminished by nitrogen fixation. Optimal pathogen growth in the nodules depends on the presence of the type-III secretion system, the structure though which bacteria introduce effector proteins into their hosts. This finding suggest that the nodule tissues mount a defense against the pathogen, which was supported by transcriptomic data. These data also show that the defense response in nodules is weaker and distinct from that of root tissue. Interestingly, the increased susceptibility of nodules to infection is counter-balanced by an unknown system that prevents dissemination of bacteria, both symbiotic and pathogenic, from the nodules and into the root system. (Summary by Mary Williams) Curr. Biol. 10.1016/j.cub.2019.11.066
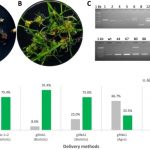
High-frequency random DNA insertions upon co-delivery of CRISPR-Cas9 ribonucleoprotein and selectable marker plasmid in rice
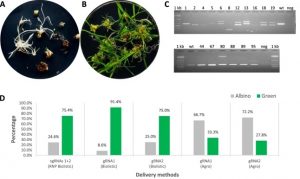 Genome editing through CRISPR/Cas9 holds so much promise for plant breeders, including potentially the ability to overcome the public’s displeasure with conventional GMOs. Still, the gene-editing RNAs and enzymes must get into the plant cell in order to work. Banakar et al. have compared three methods in terms of both the efficiency of gene editing and the introduction of unintended insertions or rearrangements at the target site. They compared delivery of CRISPR reagents (1) as plasmid DNA through Agrobacterium-mediated transformation, (2) as DNA through biolistic bombardment, and (3) as a ribonucleoprotein (RNP) complex through biolistic bombardment. Because the editing frequency has been shown to be lower when the reagents are delivered as RPN complex, they also included a DNA-encoded selectable marker. They found that both biolistic methods led to a high-frequency of insertions at the target site. The authors point out that these can easily be overlooked in PCR-based screens of the resulting plants. (Summary by Mary Williams) Sci Reports 10.1038/s41598-019-55681-y
Genome editing through CRISPR/Cas9 holds so much promise for plant breeders, including potentially the ability to overcome the public’s displeasure with conventional GMOs. Still, the gene-editing RNAs and enzymes must get into the plant cell in order to work. Banakar et al. have compared three methods in terms of both the efficiency of gene editing and the introduction of unintended insertions or rearrangements at the target site. They compared delivery of CRISPR reagents (1) as plasmid DNA through Agrobacterium-mediated transformation, (2) as DNA through biolistic bombardment, and (3) as a ribonucleoprotein (RNP) complex through biolistic bombardment. Because the editing frequency has been shown to be lower when the reagents are delivered as RPN complex, they also included a DNA-encoded selectable marker. They found that both biolistic methods led to a high-frequency of insertions at the target site. The authors point out that these can easily be overlooked in PCR-based screens of the resulting plants. (Summary by Mary Williams) Sci Reports 10.1038/s41598-019-55681-y