Plant Science Research Weekly: August 9th
Physcomitrella STEMIN transcription factor induces stem cell formation with epigenetic reprogramming
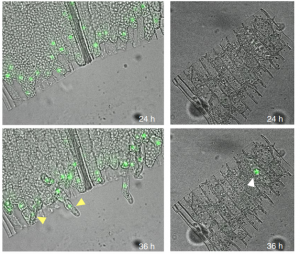 Insights into transcription factors (TFs) in non-flowering plants are increading thanks to the establishment of new bryophyte model organisms. In new work, Ishikawa et al. present STEMIN1, a Physcomitrella patens TF belonging to an uncharacterized subgroup of the AP2/ERF family. Physcomitrella undergo cell reprogramming when the leaf is cut, generating chloronema apical stem cells that grow in a filamentous form. STEMIN1 is specifically expressed in regenerating cells, and its overexpression generates ectopic stem cells. Loss-of-function of STEMIN1 and its homologs causes reduced side branching in protonemal growth. This paper provides an in-depth genome-wide analysis of downstream targets of a TF, including RNA-seq and ChiP-seq of the TF and histone marks using an inducible system. The authors show a decrease of H3K27me3 chromatin repressive mark in the promoter regions of STEMIN1-direct target genes, demonstrating how this TF regulates cell-reprogramming through an epigenetic mechanism. It is exciting to see top-notch molecular techniques being applied to early-diverging land plants. (Summary by Facundo Romani) Nature Plants 10.1038/s41477-019-0464-2.
Insights into transcription factors (TFs) in non-flowering plants are increading thanks to the establishment of new bryophyte model organisms. In new work, Ishikawa et al. present STEMIN1, a Physcomitrella patens TF belonging to an uncharacterized subgroup of the AP2/ERF family. Physcomitrella undergo cell reprogramming when the leaf is cut, generating chloronema apical stem cells that grow in a filamentous form. STEMIN1 is specifically expressed in regenerating cells, and its overexpression generates ectopic stem cells. Loss-of-function of STEMIN1 and its homologs causes reduced side branching in protonemal growth. This paper provides an in-depth genome-wide analysis of downstream targets of a TF, including RNA-seq and ChiP-seq of the TF and histone marks using an inducible system. The authors show a decrease of H3K27me3 chromatin repressive mark in the promoter regions of STEMIN1-direct target genes, demonstrating how this TF regulates cell-reprogramming through an epigenetic mechanism. It is exciting to see top-notch molecular techniques being applied to early-diverging land plants. (Summary by Facundo Romani) Nature Plants 10.1038/s41477-019-0464-2.
Arabidopsis endosperm response to paternal genome dosage is regulated by small RNA pathway
 The endosperm, a tissue that transfers nutrients from maternal tissue to the embryo, is normally triploid, with two copies of the maternal genome and one of the paternal genome. Excess paternal genomes usually leads to endosperm abnormalities and seed abortion. Previous studies have shown that the RNA polymerase IV NUCLEAR RNA POLYMERASE D1 (NRPD1) acts as an epigenetic regulator in the paternal tissue and is required for Arabidopsis interploidy seed development. Although there have been several models to depict what determines the threshold between seed lethality and viability, the mechanism has still been ambiguous. Here, Satyaki et al. used unbalanced Arabidopsis ploidy in series of genetic and genomic experiments and found that paternal RNA-directed DNA methylation (RdDM) activity genetically contributes to seed viability. Their discoveries suggest reconsideration of existing models for dosage sensitivity in endosperm and provide an important foundation for understanding hybrid incompatibilities in crops. (Summary by Jin Liao) The Plant Cell 10.1105/tpc.19.00047
The endosperm, a tissue that transfers nutrients from maternal tissue to the embryo, is normally triploid, with two copies of the maternal genome and one of the paternal genome. Excess paternal genomes usually leads to endosperm abnormalities and seed abortion. Previous studies have shown that the RNA polymerase IV NUCLEAR RNA POLYMERASE D1 (NRPD1) acts as an epigenetic regulator in the paternal tissue and is required for Arabidopsis interploidy seed development. Although there have been several models to depict what determines the threshold between seed lethality and viability, the mechanism has still been ambiguous. Here, Satyaki et al. used unbalanced Arabidopsis ploidy in series of genetic and genomic experiments and found that paternal RNA-directed DNA methylation (RdDM) activity genetically contributes to seed viability. Their discoveries suggest reconsideration of existing models for dosage sensitivity in endosperm and provide an important foundation for understanding hybrid incompatibilities in crops. (Summary by Jin Liao) The Plant Cell 10.1105/tpc.19.00047
An insulator of CLE-signaling boosts cell-differentiation in protophloem formation
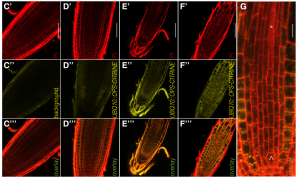 Waddington’s epigenetic landscape, a famous metaphor in developmental biology, depicts how a stem cell progresses from an undifferentiated state to a differentiated one. To some degree this metaphor can also be applied to root cell differentiation. It’s well-known that the CLE-45 peptide and its receptor BAM3 are essential for protophloem formation in Arabidopsis, but the regulation of the process of root meristem fate transition from proliferation into differentiation is unknown. Here Breda et al. found a novel mechanism in protophloem sieve element differentiation, in which a protein named OPS antagonizes CLE-45 perception by direct interference with CLE signaling in a dosage-dependent manner. The discovery reveals antagonism in the cell fate control of root meristem, which may help explain the phenotypic plasticity of plants. (Summary by Nanxun Qin) Curr. Biol. 10.1016/j.cub.2019.06.037
Waddington’s epigenetic landscape, a famous metaphor in developmental biology, depicts how a stem cell progresses from an undifferentiated state to a differentiated one. To some degree this metaphor can also be applied to root cell differentiation. It’s well-known that the CLE-45 peptide and its receptor BAM3 are essential for protophloem formation in Arabidopsis, but the regulation of the process of root meristem fate transition from proliferation into differentiation is unknown. Here Breda et al. found a novel mechanism in protophloem sieve element differentiation, in which a protein named OPS antagonizes CLE-45 perception by direct interference with CLE signaling in a dosage-dependent manner. The discovery reveals antagonism in the cell fate control of root meristem, which may help explain the phenotypic plasticity of plants. (Summary by Nanxun Qin) Curr. Biol. 10.1016/j.cub.2019.06.037
Cytokinin functions as an asymmetric and anti-gravitropic signal in lateral roots
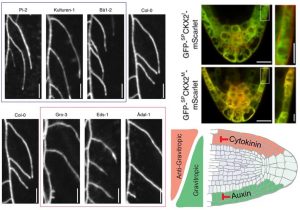 Lateral roots help plants to explore the soil and environmental conditions. Waidmann et al. took advantage of natural variants to understand lateral growth angle by analyzing gravitropic set point angle (GSA). Using Col-0 as a reference, lines that over- or under-respond (steeper or shallower root angle) were identified. From their genome-wide association study, the authors identified a SNP (T → G) that affects root angle in the CYTOKININ OXIDASE 2 (CKX2) gene; CKXs are cytokinin degrading enzymes. This SNP causes a change from isoleucine (I) to methionine (M) at the first amino acid in the mature enzyme, just after the position where the signal peptide is cleaved off. The CKX2M version has reduced secretion signaling peptide cleavage and less transport to the apoplast. Consistently, CKX2I but not CKX2M can complement a ckx2-1 loss-of-function mutant. Additionally, the authors demonstrated that asymmetric cytokinin signaling plays a crucial role as an anti-gravitropic regulator in LRs. (Summary by Arif Asraf). Nature Comms. 10.1038/s41467-019-11483-4
Lateral roots help plants to explore the soil and environmental conditions. Waidmann et al. took advantage of natural variants to understand lateral growth angle by analyzing gravitropic set point angle (GSA). Using Col-0 as a reference, lines that over- or under-respond (steeper or shallower root angle) were identified. From their genome-wide association study, the authors identified a SNP (T → G) that affects root angle in the CYTOKININ OXIDASE 2 (CKX2) gene; CKXs are cytokinin degrading enzymes. This SNP causes a change from isoleucine (I) to methionine (M) at the first amino acid in the mature enzyme, just after the position where the signal peptide is cleaved off. The CKX2M version has reduced secretion signaling peptide cleavage and less transport to the apoplast. Consistently, CKX2I but not CKX2M can complement a ckx2-1 loss-of-function mutant. Additionally, the authors demonstrated that asymmetric cytokinin signaling plays a crucial role as an anti-gravitropic regulator in LRs. (Summary by Arif Asraf). Nature Comms. 10.1038/s41467-019-11483-4
Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx
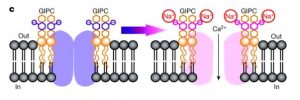 Soil salinity is one of the most important global problems that negatively affect crop productivity. Jiang et al designed a forward genetic screen in A. thaliana to identify the specific ionic response triggered by salt stress. They mutagenized plants expressing the genetically encoded Ca2+ sensor aequorin, looking at individuals with a reduced NaCl-induced cytosolic Ca2+ concentration. The screen allowed them to analyze the glucuronosyltransferase (IPUT1) for glycosyl inositol phosphorylceramide (GIPC) sphingolipids, MOCA1. The lack of MOCA1/IPUT1 activity not only compromises the salt-induced cytosolic Ca2+ increase but also affects some mechanisms, such as membrane depolarization, and makes plant Na+ sensitive. The authors highlight that MOCA1/IPUT1 is required for the production of GIPC sphingolipids which are present in the plant plasma membrane and are able to bind Na+, supporting a role of sphingolipids in the Na+ sensing in plants. The presence of these lipids let us to hypothesize that the Ca2+ channels should be in the same compartments. Last but not least genetic screens based on “imaging technologies” might represent the standard to discover new genes and proteins involved in important signaling pathways. (Summary by Francesca Resentini) Nature 10.1038/s41586-019-1449-z
Soil salinity is one of the most important global problems that negatively affect crop productivity. Jiang et al designed a forward genetic screen in A. thaliana to identify the specific ionic response triggered by salt stress. They mutagenized plants expressing the genetically encoded Ca2+ sensor aequorin, looking at individuals with a reduced NaCl-induced cytosolic Ca2+ concentration. The screen allowed them to analyze the glucuronosyltransferase (IPUT1) for glycosyl inositol phosphorylceramide (GIPC) sphingolipids, MOCA1. The lack of MOCA1/IPUT1 activity not only compromises the salt-induced cytosolic Ca2+ increase but also affects some mechanisms, such as membrane depolarization, and makes plant Na+ sensitive. The authors highlight that MOCA1/IPUT1 is required for the production of GIPC sphingolipids which are present in the plant plasma membrane and are able to bind Na+, supporting a role of sphingolipids in the Na+ sensing in plants. The presence of these lipids let us to hypothesize that the Ca2+ channels should be in the same compartments. Last but not least genetic screens based on “imaging technologies” might represent the standard to discover new genes and proteins involved in important signaling pathways. (Summary by Francesca Resentini) Nature 10.1038/s41586-019-1449-z
Male sterility in maize after transient heat stress
 The corn we eat is a seed, which is a product of fertilization. It is widely known that corn yields are highly susceptible to abiotic stress conditions that occur during the repdroductive stage of development. Begcy et al. used a metabolomic / transcriptomic approach to understand why transient heat stress (particularly during meiosis) is so detrimental to pollen viability, hence seed production. Interestingly, mild transient heat stress has big effects on several metabolites and genes involved in starch, lipid and energy synthesis. At the same time, the effects on developmental, hormone and cell wall genes, as well as those involved in the unfolded protein response (a typical response to heat) were minor. Additionally, the expression of metabolic pathway genes remained altered even after the plants were returned to optimal temperature conditions. The authors also identified a set of transcription factors that are correlated with the transcriptional response to temperature. The authors observe that these findings will useful in developing crops with greater resiliency to rising global temperatures. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.00707
The corn we eat is a seed, which is a product of fertilization. It is widely known that corn yields are highly susceptible to abiotic stress conditions that occur during the repdroductive stage of development. Begcy et al. used a metabolomic / transcriptomic approach to understand why transient heat stress (particularly during meiosis) is so detrimental to pollen viability, hence seed production. Interestingly, mild transient heat stress has big effects on several metabolites and genes involved in starch, lipid and energy synthesis. At the same time, the effects on developmental, hormone and cell wall genes, as well as those involved in the unfolded protein response (a typical response to heat) were minor. Additionally, the expression of metabolic pathway genes remained altered even after the plants were returned to optimal temperature conditions. The authors also identified a set of transcription factors that are correlated with the transcriptional response to temperature. The authors observe that these findings will useful in developing crops with greater resiliency to rising global temperatures. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.00707
TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity
 Identification of protein- protein interactions helps understand the signaling cascade in multiple biological process including development, biotic and abiotic stress. Previously, a proximity-based labelling approach referred to as BioID had been used to identify proteins in close proximity to a protein of interest by labelling them with biotin. In this paper, Zhang et al. optimized TURBOID to identify proteins that interact with the N NLR (Nucleotide binding Leucine rich repeat Receptor) during biotic stress. The N receptor was triggered with p50 elicitor, and proteins in close proximity were labeled and identified by mass spectrometry. UBR7, one of the identified proteins, was shown to negatively regulate the receptor and alter the immune response in tobacco. Thus the TURBOID tool enabled identification of potential interactors that are crucial to receptor-mediated plant defense. (Summary by Suresh Damodaran) Nature Comms. 10.1038/s41467-019-11202-z.
Identification of protein- protein interactions helps understand the signaling cascade in multiple biological process including development, biotic and abiotic stress. Previously, a proximity-based labelling approach referred to as BioID had been used to identify proteins in close proximity to a protein of interest by labelling them with biotin. In this paper, Zhang et al. optimized TURBOID to identify proteins that interact with the N NLR (Nucleotide binding Leucine rich repeat Receptor) during biotic stress. The N receptor was triggered with p50 elicitor, and proteins in close proximity were labeled and identified by mass spectrometry. UBR7, one of the identified proteins, was shown to negatively regulate the receptor and alter the immune response in tobacco. Thus the TURBOID tool enabled identification of potential interactors that are crucial to receptor-mediated plant defense. (Summary by Suresh Damodaran) Nature Comms. 10.1038/s41467-019-11202-z.



