Plant Science Research Weekly: August 21, 2020
Stimulating photosynthetic processes increases productivity and water-use efficiency in the field
 Yield potential in crops is determined by the efficiency of photosynthetic rates, which is a critical target for improvement. Previous studies have shown that photosynthetic carbon assimilation and plant biomass can be improved by the stimulation of either regeneration of RuBP (the five-carbon sugar that CO2 attaches to) or the electron transport chain. López-Calcagno et al. tested whether combining stimulation from both these processes could enhance photosynthesis and yield more as compared to stimulation of only one process. Two strategies were used to increase RuBP regeneration: expressing a cyanobacterial bifunctional enzyme fructose-1,6-bisphosphatase/sedoheptulose-1,7-bisphosphatase (FBP/SBPase) in SB plants, or overexpression of the plant enzyme sedoheptulose-1,7-bisphosphatase (SBPase) in S plants. Electron transport was enhanced by expression of the red algal protein cytochrome c6 (C6). The simultaneous stimulation of both processes in either SBC6 or SC6 plants had a greater impact on photosynthesis in comparison to the single manipulations (SB, S, or C6) in all plants analyzed. The double transgenics also presented increased biomass and yield under greenhouse and field conditions. Interestingly, in the field trial, stomatal conductance (gs) in the SBC6 plants was significantly lower, resulting in a higher intrinsic water-use efficiency (iWUE) for SBC6 plants. This genetic engineering approach has the potential to be further explored as a tool to enhance yield and reduce water consumption. (Summary by Camila Ribeiro @camicribeiro86) Nature Plants 10.1038/s41477-020-0740-1
Yield potential in crops is determined by the efficiency of photosynthetic rates, which is a critical target for improvement. Previous studies have shown that photosynthetic carbon assimilation and plant biomass can be improved by the stimulation of either regeneration of RuBP (the five-carbon sugar that CO2 attaches to) or the electron transport chain. López-Calcagno et al. tested whether combining stimulation from both these processes could enhance photosynthesis and yield more as compared to stimulation of only one process. Two strategies were used to increase RuBP regeneration: expressing a cyanobacterial bifunctional enzyme fructose-1,6-bisphosphatase/sedoheptulose-1,7-bisphosphatase (FBP/SBPase) in SB plants, or overexpression of the plant enzyme sedoheptulose-1,7-bisphosphatase (SBPase) in S plants. Electron transport was enhanced by expression of the red algal protein cytochrome c6 (C6). The simultaneous stimulation of both processes in either SBC6 or SC6 plants had a greater impact on photosynthesis in comparison to the single manipulations (SB, S, or C6) in all plants analyzed. The double transgenics also presented increased biomass and yield under greenhouse and field conditions. Interestingly, in the field trial, stomatal conductance (gs) in the SBC6 plants was significantly lower, resulting in a higher intrinsic water-use efficiency (iWUE) for SBC6 plants. This genetic engineering approach has the potential to be further explored as a tool to enhance yield and reduce water consumption. (Summary by Camila Ribeiro @camicribeiro86) Nature Plants 10.1038/s41477-020-0740-1
BonnMu: a sequence-indexed resource of transposon-induced maize mutations for functional genomics studies
 Loss-of-function mutants have been invaluable tools in the geneticist’s toolbox for more than a century. More recently, libraries have been developed of knockout insertion mutants at known sites. Here, a new Mu-transposon insertion sequence-indexed maize resource is described. This new collection, BonnMu (developed in Bonn, Germany) complements the North American UniformMu and the Asian ChinaMu resources. A comparison of the BonnMu data from the B73 line and the UniformMu from the W22 line shows an abundance of conserved insertion hotspots between different genetic backgrounds. Furthermore, most insertions from both datasets were close to the transcription start site, which corresponds to open chromatin. The BonnMu collection provides easy-to-view images of segregating F2 populations that can be accessed from the genome browser, https://www.maizegdb.org/gbrowse. From the genome browser view, click “Select track” and under “Insertions” select “BonnMu insertions”. Go back to the browser and the insertion sites become visible in the locus. Clicking on the insertion site links to the Mu insertion identifiers (e.g., BonnMu0000812) and the respective F2–Mu-seq family (e.g., BonnMu-2-A-0596) with an image of its phenotype 10 days after germination. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1104/pp.20.00478
Loss-of-function mutants have been invaluable tools in the geneticist’s toolbox for more than a century. More recently, libraries have been developed of knockout insertion mutants at known sites. Here, a new Mu-transposon insertion sequence-indexed maize resource is described. This new collection, BonnMu (developed in Bonn, Germany) complements the North American UniformMu and the Asian ChinaMu resources. A comparison of the BonnMu data from the B73 line and the UniformMu from the W22 line shows an abundance of conserved insertion hotspots between different genetic backgrounds. Furthermore, most insertions from both datasets were close to the transcription start site, which corresponds to open chromatin. The BonnMu collection provides easy-to-view images of segregating F2 populations that can be accessed from the genome browser, https://www.maizegdb.org/gbrowse. From the genome browser view, click “Select track” and under “Insertions” select “BonnMu insertions”. Go back to the browser and the insertion sites become visible in the locus. Clicking on the insertion site links to the Mu insertion identifiers (e.g., BonnMu0000812) and the respective F2–Mu-seq family (e.g., BonnMu-2-A-0596) with an image of its phenotype 10 days after germination. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1104/pp.20.00478
A constitutively monomeric UVR8 photoreceptor allele confers enhanced UV-B photomorphogenesis
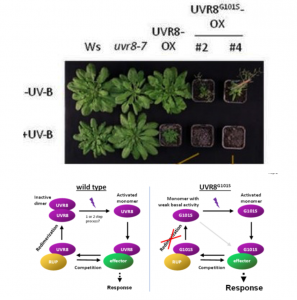 In response to UV-B light, the plant UV-B photoreceptor UV RESISTANCE LOCUS 8 (UVR8) monomerizes and activates, inducing UV-B acclimation and survival. Re-dimerization by REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 (RUP1) and RUP2 inactivates UVR8. In order to further clarify UVR8 pathway regulation, Podolec et al. analyzed Arabidopsis mutants with an aberrant hypocotyl phenotype when grown for 4 days under weak white light supplemented with UV-B. They discovered a novel hypersensitive uvr8 mutant allele, uvr8-17D, that harbors the UVR8G101S protein variant which has a single glycine-101 to serine amino acid change. Seedlings of uvr8-17D showed enhanced UV-B acclimation and stress tolerance, which were associated with UVR8’s monomeric conformation in vivo, suggesting that re-dimerization by RUP1 and RUP2 is impaired and that UVR8G101S remains active much longer than wild-type UVR8. This novel allele represents an exciting possibility to generate UV-B-hypersensitive phenotypes, including enhanced acclimation and UV-B tolerance, in other species by a single nucleotide substitution. (Summary by Elisandra Pradella @Elisandra_MP) bioRxiv 10.1101/2020.08.02.233007
In response to UV-B light, the plant UV-B photoreceptor UV RESISTANCE LOCUS 8 (UVR8) monomerizes and activates, inducing UV-B acclimation and survival. Re-dimerization by REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 (RUP1) and RUP2 inactivates UVR8. In order to further clarify UVR8 pathway regulation, Podolec et al. analyzed Arabidopsis mutants with an aberrant hypocotyl phenotype when grown for 4 days under weak white light supplemented with UV-B. They discovered a novel hypersensitive uvr8 mutant allele, uvr8-17D, that harbors the UVR8G101S protein variant which has a single glycine-101 to serine amino acid change. Seedlings of uvr8-17D showed enhanced UV-B acclimation and stress tolerance, which were associated with UVR8’s monomeric conformation in vivo, suggesting that re-dimerization by RUP1 and RUP2 is impaired and that UVR8G101S remains active much longer than wild-type UVR8. This novel allele represents an exciting possibility to generate UV-B-hypersensitive phenotypes, including enhanced acclimation and UV-B tolerance, in other species by a single nucleotide substitution. (Summary by Elisandra Pradella @Elisandra_MP) bioRxiv 10.1101/2020.08.02.233007
Going my own way: Ethylene-independent ACC signaling in pollen tube attraction
 1-Aminocyclopropane-1-carboxylic acid (ACC) is the precursor of the phytohormone ethylene. Here, Mou and co-workers show ACC to act independent of ethylene in the ovule to attract growing pollen tubes. Mutants impaired in ACC biosynthesis had less fertilization and seed setting events. Fertilization and seed set were both rescued by external application of ACC but not ethylene. Visualization of pollen tube growth showed that very few pollen tubes precisely targeted the ovule of mutant flowers. This indicates that ovules of mutants may be unable to attract pollen tubes, an idea that is supported by the finding that LURE1, a peptide that acts as pollen tube attractant, is not properly secreted in the mutants. In neurons, ACC has been shown to act as an agonist for glutamate receptor-like (GLR) calcium channels. Here, the authors found that ACC could also induce Ca2+ transport in ovules. This work underlines the limitations of using ACC as an ethylene supplement and proposes a novel role for ACC in plant Ca2+ signaling. (Summary by Pavithran Narayanan @pavi_narayanan) Nature Comms. 10.1038/s41467-020-17819-9
1-Aminocyclopropane-1-carboxylic acid (ACC) is the precursor of the phytohormone ethylene. Here, Mou and co-workers show ACC to act independent of ethylene in the ovule to attract growing pollen tubes. Mutants impaired in ACC biosynthesis had less fertilization and seed setting events. Fertilization and seed set were both rescued by external application of ACC but not ethylene. Visualization of pollen tube growth showed that very few pollen tubes precisely targeted the ovule of mutant flowers. This indicates that ovules of mutants may be unable to attract pollen tubes, an idea that is supported by the finding that LURE1, a peptide that acts as pollen tube attractant, is not properly secreted in the mutants. In neurons, ACC has been shown to act as an agonist for glutamate receptor-like (GLR) calcium channels. Here, the authors found that ACC could also induce Ca2+ transport in ovules. This work underlines the limitations of using ACC as an ethylene supplement and proposes a novel role for ACC in plant Ca2+ signaling. (Summary by Pavithran Narayanan @pavi_narayanan) Nature Comms. 10.1038/s41467-020-17819-9
Hydration-dependent phase separation of a prion-like protein regulates seed germination during water stress
 Plant seeds can remain dormant for several years under stress conditions and subsequently germinate once favorable conditions return. Even though this phenomenon has been known for many years, what keeps the seed from germinating during the unfavorable conditions, especially during water deficit conditions, is not known. In this paper, Dorone et al. identified an intrinsically disordered protein (IDP) with a prion-like domain that they named FLOE1 (floe meaning a “sheet of floating ice”) that prevents seed germination during water stress conditions. Phylogenetic analysis shows that FLOE proteins are conserved in all green plants and predates seed evolution. The authors showed that FLOE1 distributes uniformly during dry conditions and phase separates to forms condensates during favorable conditions. Through deletion of various domains, the authors generated variant proteins with different condensation and gelling properties, which revealed the key role of FLOE1 in controlling germination. Loss-of-function mutants germinate (and die) under unfavorable conditions, and expression of the ΔDS protein that forms larger and more stable condensates than WT also showed greatly enhanced germination under salt-stress conditions. The authors suggest that FLOE1 acts upstream of key germination pathways, and that it “might act as a molecular glue helping to stabilize the desiccated glassy state” and prevent germination. Thus, this paper provides molecular insights into seed dormancy during water stress conditions and has agronomical implications in crop improvement. (Summary by Vijaya Batthula @Vijaya_Batthula) bioRxiv 10.1101/2020.08.07.242172
Plant seeds can remain dormant for several years under stress conditions and subsequently germinate once favorable conditions return. Even though this phenomenon has been known for many years, what keeps the seed from germinating during the unfavorable conditions, especially during water deficit conditions, is not known. In this paper, Dorone et al. identified an intrinsically disordered protein (IDP) with a prion-like domain that they named FLOE1 (floe meaning a “sheet of floating ice”) that prevents seed germination during water stress conditions. Phylogenetic analysis shows that FLOE proteins are conserved in all green plants and predates seed evolution. The authors showed that FLOE1 distributes uniformly during dry conditions and phase separates to forms condensates during favorable conditions. Through deletion of various domains, the authors generated variant proteins with different condensation and gelling properties, which revealed the key role of FLOE1 in controlling germination. Loss-of-function mutants germinate (and die) under unfavorable conditions, and expression of the ΔDS protein that forms larger and more stable condensates than WT also showed greatly enhanced germination under salt-stress conditions. The authors suggest that FLOE1 acts upstream of key germination pathways, and that it “might act as a molecular glue helping to stabilize the desiccated glassy state” and prevent germination. Thus, this paper provides molecular insights into seed dormancy during water stress conditions and has agronomical implications in crop improvement. (Summary by Vijaya Batthula @Vijaya_Batthula) bioRxiv 10.1101/2020.08.07.242172
From one cell to many: Morphogenetic field of lateral root founder cells in Arabidopsis thaliana is built by gradual recruitment
 Because plant cells don’t usually move within an organ, it is possible to trace the developmental program cell by cell, through a technique called clonal analysis. If a single cell can be labeled in some heritable way, then all its progeny will also carry this label. Torres-Martínez et al. randomly induced expression of YPF in individual cells to investigate lateral root development in Arabidopsis. Lateral roots initiate from xylem-adjacent cells of the pericycle, and lateral root numbers vary depending on environmental conditions. The authors found that in most cases a single pericycle cell initiates the process of lateral root development and serves as the “founder cell”. Through an auxin-dependent process, the initial founder cell swiftly recruits adjacent cells also to become founder cells and contribute to the formation of the nascent lateral root, forming what is described as a morphogenetic field. The authors also note the distinction between founder cells, with progeny that all become part of the new lateral root, and stem cells, in which at least one of the progeny retains the original cell identity. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2006387117
Because plant cells don’t usually move within an organ, it is possible to trace the developmental program cell by cell, through a technique called clonal analysis. If a single cell can be labeled in some heritable way, then all its progeny will also carry this label. Torres-Martínez et al. randomly induced expression of YPF in individual cells to investigate lateral root development in Arabidopsis. Lateral roots initiate from xylem-adjacent cells of the pericycle, and lateral root numbers vary depending on environmental conditions. The authors found that in most cases a single pericycle cell initiates the process of lateral root development and serves as the “founder cell”. Through an auxin-dependent process, the initial founder cell swiftly recruits adjacent cells also to become founder cells and contribute to the formation of the nascent lateral root, forming what is described as a morphogenetic field. The authors also note the distinction between founder cells, with progeny that all become part of the new lateral root, and stem cells, in which at least one of the progeny retains the original cell identity. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2006387117
Friend vs. foe: molecular insight towards microbial recognition and specificity in legume signaling
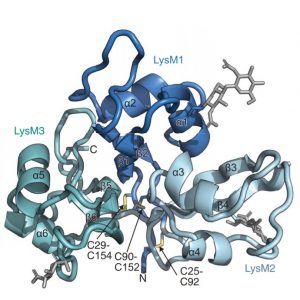 Ecosystems are founded by relationships between organisms, such as the mutualism between plants and microbes. An example is the agriculturally important symbiosis between legumes and nitrogen-fixing bacteria that reside in nodules of legumes. Legumes must be able to differentiate, through undetermined mechanisms, between symbiotic and pathogenic microbes. Boszoki et al. set out to elucidate the molecular players used by a model legume, Lotus japonicus, to identify microbes and initiate the proper response. NFR1 and CERK6 are receptor proteins of L. japonicus with similar structures and previously-demonstrated necessary function for nodule organogenesis and chitin immune response, respectively. Through constructing chimeric versions of each protein by swapping residues/domains from either NFR1 or CERK6, expressing these chimeras in L. japonicus lines, and comparing the resulting plants’ abilities to form nodules (a readout of recognizing nodulation factors from symbiotic bacteria, via NFR1 signaling) or induce an immune response (a readout of recognizing chitin from pathogenic fungi, via CERK6 signaling), the authors deduced that differentiation between symbiotic or pathogenic microbes is derived from the extracellular domain, composed of three lysin motif (LysM) domains. Sequence analysis and experiments honing into these motifs demonstrated that two specific regions within the first LysM (LysM1) domain were required for specific signaling response. The authors further demonstrated the important applicability of these conclusions by engineering a chimeric receptor using the two aforementioned regions of the first LysM1 domain from NFR1 within a CERK6 ectodomain background and demonstrating its ability to bind nodulation factors and induce nodule formation. These findings are imperative for decoding the molecular languages of plants and microbes so that we may engineer more adaptable crops for a changing environment. (Summary by Benjamin Jin) Science 10.1126/science.abb3377
Ecosystems are founded by relationships between organisms, such as the mutualism between plants and microbes. An example is the agriculturally important symbiosis between legumes and nitrogen-fixing bacteria that reside in nodules of legumes. Legumes must be able to differentiate, through undetermined mechanisms, between symbiotic and pathogenic microbes. Boszoki et al. set out to elucidate the molecular players used by a model legume, Lotus japonicus, to identify microbes and initiate the proper response. NFR1 and CERK6 are receptor proteins of L. japonicus with similar structures and previously-demonstrated necessary function for nodule organogenesis and chitin immune response, respectively. Through constructing chimeric versions of each protein by swapping residues/domains from either NFR1 or CERK6, expressing these chimeras in L. japonicus lines, and comparing the resulting plants’ abilities to form nodules (a readout of recognizing nodulation factors from symbiotic bacteria, via NFR1 signaling) or induce an immune response (a readout of recognizing chitin from pathogenic fungi, via CERK6 signaling), the authors deduced that differentiation between symbiotic or pathogenic microbes is derived from the extracellular domain, composed of three lysin motif (LysM) domains. Sequence analysis and experiments honing into these motifs demonstrated that two specific regions within the first LysM (LysM1) domain were required for specific signaling response. The authors further demonstrated the important applicability of these conclusions by engineering a chimeric receptor using the two aforementioned regions of the first LysM1 domain from NFR1 within a CERK6 ectodomain background and demonstrating its ability to bind nodulation factors and induce nodule formation. These findings are imperative for decoding the molecular languages of plants and microbes so that we may engineer more adaptable crops for a changing environment. (Summary by Benjamin Jin) Science 10.1126/science.abb3377


