Plant Science Research Weekly: August 12, 2022
Science special issue on Grasses
 “Grassy biomes—from the steppes of Mongolia to the savannas of Tanzania— are predicted to be the ecosystems hardest hit by the ongoing climate and land use crises,” begins a Perspective by Strömberg and Staver in the August 4, 2022 Science special issue on Grasses that raises awareness of the unrecognized value of grasses and grasslands. Grasses, particularly rice, wheat, and corn, provide 50% of calories in the human diet (more when you add in sugar cane and barley for beer) and grasslands provide habitat for a great diversity of animals. The fascinating collection has review articles about the developmental and genetic diversity of grasses, emphasizing traits that have been selected to increase yield including nonshattering traits and inflorescence architecture; evolutionary history of grassy biomes; contributions of grasslands to soil carbon sequestration; challenges to “old growth” grassland restoration; and a plea to recognize the ecological roles of seagrass in global conservation efforts. Beautifully illustrated, this is an excellent resource to share with friends and students. (Summary by Mary Williams @PlantTeaching) Science https://www.science.org/toc/science/377/6606.
“Grassy biomes—from the steppes of Mongolia to the savannas of Tanzania— are predicted to be the ecosystems hardest hit by the ongoing climate and land use crises,” begins a Perspective by Strömberg and Staver in the August 4, 2022 Science special issue on Grasses that raises awareness of the unrecognized value of grasses and grasslands. Grasses, particularly rice, wheat, and corn, provide 50% of calories in the human diet (more when you add in sugar cane and barley for beer) and grasslands provide habitat for a great diversity of animals. The fascinating collection has review articles about the developmental and genetic diversity of grasses, emphasizing traits that have been selected to increase yield including nonshattering traits and inflorescence architecture; evolutionary history of grassy biomes; contributions of grasslands to soil carbon sequestration; challenges to “old growth” grassland restoration; and a plea to recognize the ecological roles of seagrass in global conservation efforts. Beautifully illustrated, this is an excellent resource to share with friends and students. (Summary by Mary Williams @PlantTeaching) Science https://www.science.org/toc/science/377/6606.
Review: Deep origin and gradual evolution of transporting tissues: Perspectives from across the land plants
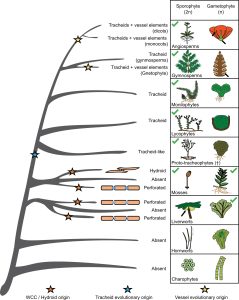 The development of vascular plants (tracheophytes) was a major innovation during the evolution of land plants. The classic view is that the evolution of transporting tissues is distinct to tracheophytes, but Woudenberg and Renema et al. discuss an alternative hypothesis where this was a gradual process that is already seen in bryophytes. The authors propose using the terms water-conducting cells (WCCs) and food-conducting cells (FCCs) to encompass these tissues with similar properties across bryophytes and tracheophytes, comparing bryophyte hydroids and leptoids with tracheophyte tracheids and sieve elements. They emphasise the importance of fossil records in elucidating the deep evolutionary origin of these tissues, discussing the hypothesis for a common origin of FCCs in bryophytes and tracheophytes. The incomplete fossil records, especially for bryophytes, still leave many questions unexplored. Although molecular insights for bryophyte transport tissue are also lacking, work from model systems shows promise in studying some highly conserved proteins and processes in vasculature development. In WCC development, auxin and cytokinin signalling are conserved but their exact roles may differ in bryophytes compared to Arabidopsis. Also, lignin in the cell wall is unique to tracheophytes, but enzymes involved in making the precursors for lignin are present in bryophytes; they might perform other roles or are involved in the synthesis of lignin-like components. By combining advances in studying the fossil record and identifying conserved proteins with distinct roles in tracheophytes and bryophytes, we are closer to gaining a comprehensive understanding of transport tissue evolution in land plants. (Summary by Jiawen Chen) Plant Physiol. 10.1093/plphys/kiac304
The development of vascular plants (tracheophytes) was a major innovation during the evolution of land plants. The classic view is that the evolution of transporting tissues is distinct to tracheophytes, but Woudenberg and Renema et al. discuss an alternative hypothesis where this was a gradual process that is already seen in bryophytes. The authors propose using the terms water-conducting cells (WCCs) and food-conducting cells (FCCs) to encompass these tissues with similar properties across bryophytes and tracheophytes, comparing bryophyte hydroids and leptoids with tracheophyte tracheids and sieve elements. They emphasise the importance of fossil records in elucidating the deep evolutionary origin of these tissues, discussing the hypothesis for a common origin of FCCs in bryophytes and tracheophytes. The incomplete fossil records, especially for bryophytes, still leave many questions unexplored. Although molecular insights for bryophyte transport tissue are also lacking, work from model systems shows promise in studying some highly conserved proteins and processes in vasculature development. In WCC development, auxin and cytokinin signalling are conserved but their exact roles may differ in bryophytes compared to Arabidopsis. Also, lignin in the cell wall is unique to tracheophytes, but enzymes involved in making the precursors for lignin are present in bryophytes; they might perform other roles or are involved in the synthesis of lignin-like components. By combining advances in studying the fossil record and identifying conserved proteins with distinct roles in tracheophytes and bryophytes, we are closer to gaining a comprehensive understanding of transport tissue evolution in land plants. (Summary by Jiawen Chen) Plant Physiol. 10.1093/plphys/kiac304
Review: Plasma membrane-to-organelle communication in plant stress signaling
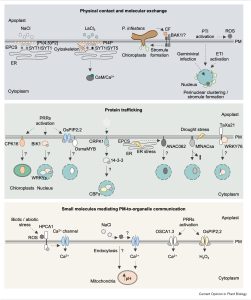 The plasma membrane (PM) is a critical interface between the cell and its environment and serves crucial sensing and transducing roles. This timely review by Medina-Puche and Lozano-Durán updates exciting new developments in understanding communication between the PM and intracellular organelles, focusing on abiotic and biotic stresses. The review sorts these interactions into three types: physical contact and molecular exchange (for example direct contacts between the endoplasmic reticulum and plasma membrane); protein trafficking (e.g., internalization of membrane-bound receptors); and small molecules mediating communication (e.g., Ca2+, H+, ROS). Each of these types is illustrated with several examples, to highlight the commonalities and distinctions. This is an excellent, thought-provoking review that highlights the many ways that information flows from out to in. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2022.102269
The plasma membrane (PM) is a critical interface between the cell and its environment and serves crucial sensing and transducing roles. This timely review by Medina-Puche and Lozano-Durán updates exciting new developments in understanding communication between the PM and intracellular organelles, focusing on abiotic and biotic stresses. The review sorts these interactions into three types: physical contact and molecular exchange (for example direct contacts between the endoplasmic reticulum and plasma membrane); protein trafficking (e.g., internalization of membrane-bound receptors); and small molecules mediating communication (e.g., Ca2+, H+, ROS). Each of these types is illustrated with several examples, to highlight the commonalities and distinctions. This is an excellent, thought-provoking review that highlights the many ways that information flows from out to in. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2022.102269
Spatial resolution of an integrated C4+CAM photosynthetic metabolism
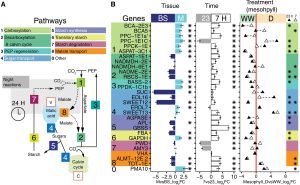 C4 and CAM are both photosynthetic strategies that concentrate CO2 upstream of RuBisCO, somewhat uncoupling photosynthetic carbon fixation from transpirational water loss. Both strategies use the enzyme PEP-carboxylase to produce an organic acid, which can then be decarboxylated to provide CO2 to RuBisCO. In the C4 system, PEP-carboxylase operates in mesophyll cells and RuBisCO in bundle sheath cells, with the organic acid shuttling between them as a CO2 carrier. Rather than this physical separation, CAM plants separate their carboxylases by time; CO2 uptake occurs at night when transpiration is lower, it is stored as organic acids in the vacuole, and then released by decarboxylation in daylight when RuBisCO is active. Moreno-Villena et al. explored the carbon-concentrating mechanisms of Portulaca oleracea, a C4 species that also carries out CAM under drought conditions. They carried out RNA sequencing on isolated mesophyll and bundle sheath cells from well-watered and droughted plants. Their first surprise was that genes involved in both C4 and CAM were expressed in the same bundle sheath cells, arguing against a previous assumption that these reactions took place in spatial isolation. The second finding was that the two pathways are actually linked. This finding comes from a metabolic model as well as flux balance analysis. Under drought conditions, CAM-produced malate feeds into the C4 decarboxylation cycle and final CO2 assimilation. The authors then speculate on why this efficient coupling of C4 and CAM is apparently rare, suggesting that Portulaca was most likely an ancestrally C3+CAM species that evolved C4. They also observe that as fast-growing, self-compatible plants, Portulaca is amenable to development as a model system, and that further study could lead to the incorporation of CAM into C4 crops, thereby increasing their drought tolerance without sacrificing productivity. (Summary by Mary Williams @PlantTeaching) Sci. Advances 10.1126/sciadv.abn2349
C4 and CAM are both photosynthetic strategies that concentrate CO2 upstream of RuBisCO, somewhat uncoupling photosynthetic carbon fixation from transpirational water loss. Both strategies use the enzyme PEP-carboxylase to produce an organic acid, which can then be decarboxylated to provide CO2 to RuBisCO. In the C4 system, PEP-carboxylase operates in mesophyll cells and RuBisCO in bundle sheath cells, with the organic acid shuttling between them as a CO2 carrier. Rather than this physical separation, CAM plants separate their carboxylases by time; CO2 uptake occurs at night when transpiration is lower, it is stored as organic acids in the vacuole, and then released by decarboxylation in daylight when RuBisCO is active. Moreno-Villena et al. explored the carbon-concentrating mechanisms of Portulaca oleracea, a C4 species that also carries out CAM under drought conditions. They carried out RNA sequencing on isolated mesophyll and bundle sheath cells from well-watered and droughted plants. Their first surprise was that genes involved in both C4 and CAM were expressed in the same bundle sheath cells, arguing against a previous assumption that these reactions took place in spatial isolation. The second finding was that the two pathways are actually linked. This finding comes from a metabolic model as well as flux balance analysis. Under drought conditions, CAM-produced malate feeds into the C4 decarboxylation cycle and final CO2 assimilation. The authors then speculate on why this efficient coupling of C4 and CAM is apparently rare, suggesting that Portulaca was most likely an ancestrally C3+CAM species that evolved C4. They also observe that as fast-growing, self-compatible plants, Portulaca is amenable to development as a model system, and that further study could lead to the incorporation of CAM into C4 crops, thereby increasing their drought tolerance without sacrificing productivity. (Summary by Mary Williams @PlantTeaching) Sci. Advances 10.1126/sciadv.abn2349
DREB1C is a regulator of nitrogen use efficiency and flowering time in rice
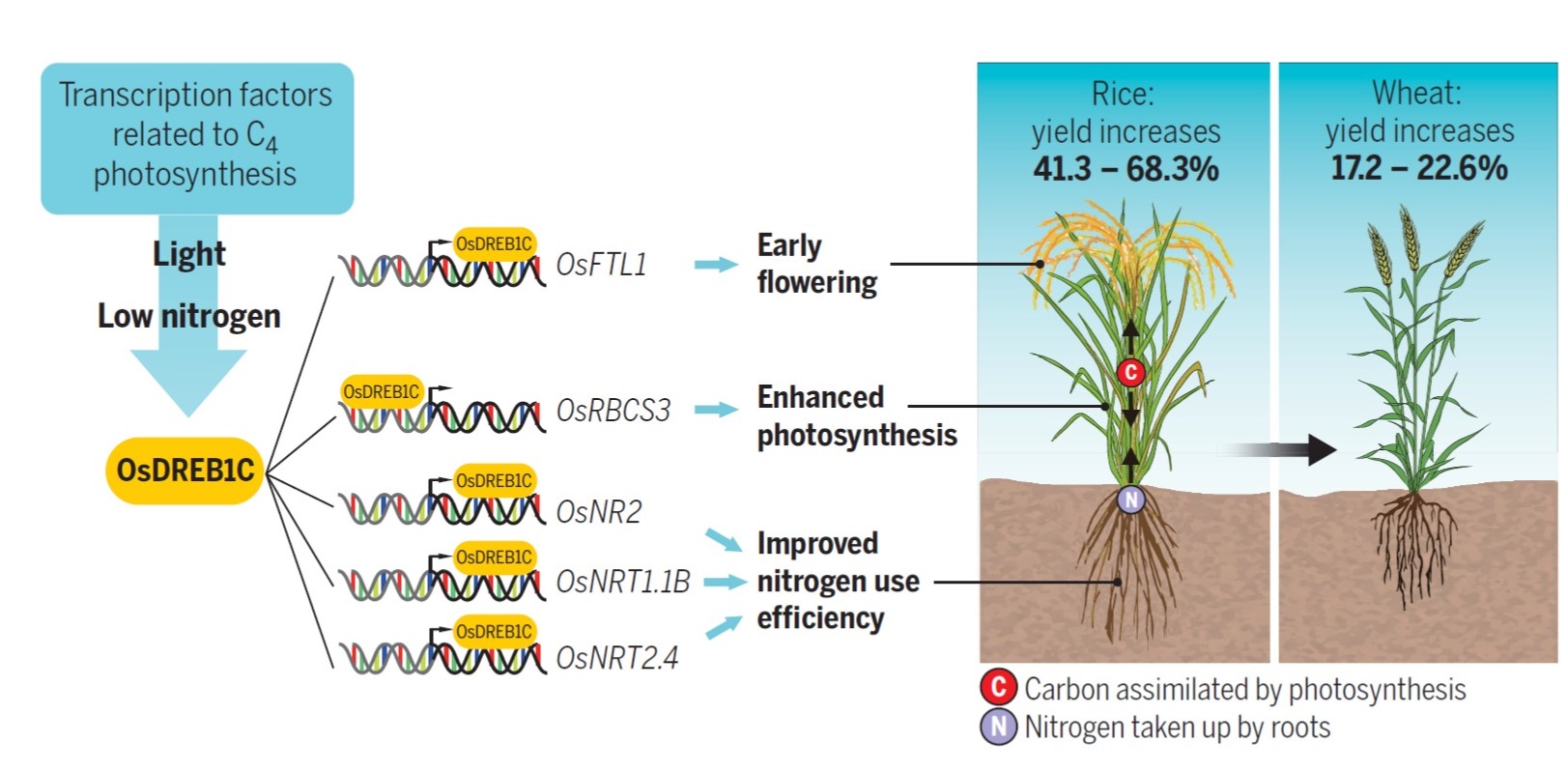 To improve the profitability and sustainability of agriculture, there has been an extensive quest to identify regulators that boost nitrogen use efficiency (NUE) without compromising yield. Wei et al. identified a DREB family transcription factor based on RNAseq analysis of rice plants exposed to low N. Among the differentially regulated transcription factors, DREB1C showed a significant induction by both low N and high light conditions. DREB1C overexpression (OE) boosted rice and wheat grain yield, while in Arabidopsis, a significant increase in total biomass was recorded. In rice, DREB1C overexpression improved early seedling vigour, abundance of photosynthetic pigments, RuBisCO content and rate of photosynthesis. There also was a significant improvement in 15N uptake and nitrogen use efficienty. DREB1C regulates the expression of several important growth-related genes including nitrate transporters (NRT1.1B, NRT2.4), nitrate reductase (NR2), flowering regulator (FTL3) and RuBisCO (RBCS3) by binding in the intragenic region or promoter regions. The DREB1C-OE plants displayed a higher harvest index and increased remobilisation of N and C to sink organs and showed a significant reduction in flowering time and crop duration due to modulation of the flowering gene (FTL3). This is an important proof-of-concept paper showing how a single transcription factor can have broad beneficial effects in rice (multiple cultivars), wheat, and Arabidopsis. Interestingly, the paper has been extensively discussed post-publication, largely because the potential for improving yields in the field in elite varieties remains to be confirmed (https://twitter.com/MerKhaiBurch/status/1553072144426405889?s=20&t=FamfyXjiP2YIloHYZKTp8w). We look forward to further tests of the effects of DREB1C overexpression in larger-scale yield trials. (Summary by Lekshmy Sathee @lekshmysnair) Science 10.1126/science.abi8455
To improve the profitability and sustainability of agriculture, there has been an extensive quest to identify regulators that boost nitrogen use efficiency (NUE) without compromising yield. Wei et al. identified a DREB family transcription factor based on RNAseq analysis of rice plants exposed to low N. Among the differentially regulated transcription factors, DREB1C showed a significant induction by both low N and high light conditions. DREB1C overexpression (OE) boosted rice and wheat grain yield, while in Arabidopsis, a significant increase in total biomass was recorded. In rice, DREB1C overexpression improved early seedling vigour, abundance of photosynthetic pigments, RuBisCO content and rate of photosynthesis. There also was a significant improvement in 15N uptake and nitrogen use efficienty. DREB1C regulates the expression of several important growth-related genes including nitrate transporters (NRT1.1B, NRT2.4), nitrate reductase (NR2), flowering regulator (FTL3) and RuBisCO (RBCS3) by binding in the intragenic region or promoter regions. The DREB1C-OE plants displayed a higher harvest index and increased remobilisation of N and C to sink organs and showed a significant reduction in flowering time and crop duration due to modulation of the flowering gene (FTL3). This is an important proof-of-concept paper showing how a single transcription factor can have broad beneficial effects in rice (multiple cultivars), wheat, and Arabidopsis. Interestingly, the paper has been extensively discussed post-publication, largely because the potential for improving yields in the field in elite varieties remains to be confirmed (https://twitter.com/MerKhaiBurch/status/1553072144426405889?s=20&t=FamfyXjiP2YIloHYZKTp8w). We look forward to further tests of the effects of DREB1C overexpression in larger-scale yield trials. (Summary by Lekshmy Sathee @lekshmysnair) Science 10.1126/science.abi8455
Increasing the resilience of plant immunity to a warming climate
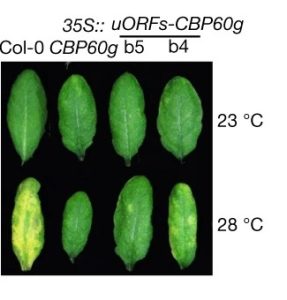 Plants demonstrate increased susceptibility to pathogens upon exposure to heat stress, apparently due to suppressed salicylic acid (SA) accumulation and subsequently decreased effector-triggered immunity. How exactly does heat stress cause this suppression, and how can we take advantage of genetics to increase plant resilience? Kim et al. discovered that heat stress acts on SA immunity pathways independent of established thermosensing mechanisms (specifically, phyB and ELF3). Rather, heat stress inhibits the formation of phase-separating condensates composed of GBPL3, a positive regulator of SA function that contains an intrinsically disordered region within its structure. GBPL3 condensates are necessary for proper transcription of CBP60g, which codes for a broad regulator of immune responses that binds to the promoter of the SA biosynthesis gene ICS1. Constitutive overexpression of CBP60g restored SA accumulation and associated immunity under heat stress, demonstrating a plant with increased resilience in the face of increasing temperatures. This genotype, however, presented a late flowering phenotype, in agreement with the growth-defense trade off; the authors thus fine-tuned their engineering by linking overexpressed CBP60g with uORFsTBF1, limiting CBP60g overexpression to pathogen infection events. This uORFsTBF1–CBP60g plant demonstrated increased resilience alongside wild-type development. Interestingly, overexpression of either GBPL3 or ICS1 did not restore immunity under heat stress, the former due to heat stress specifically inhibiting GBPL3 condensate formation, and the latter likely due to the broader role that CBP60g plays in plant immune responses. Altogether, this work highlights the power of genetic engineering to develop plants that can withstand climate change, and the importance of dissecting how physiological responses operate down to a molecular level of detail. (Summary by Benjamin Jin) Nature, 10.1038/s41586-022-04902-y
Plants demonstrate increased susceptibility to pathogens upon exposure to heat stress, apparently due to suppressed salicylic acid (SA) accumulation and subsequently decreased effector-triggered immunity. How exactly does heat stress cause this suppression, and how can we take advantage of genetics to increase plant resilience? Kim et al. discovered that heat stress acts on SA immunity pathways independent of established thermosensing mechanisms (specifically, phyB and ELF3). Rather, heat stress inhibits the formation of phase-separating condensates composed of GBPL3, a positive regulator of SA function that contains an intrinsically disordered region within its structure. GBPL3 condensates are necessary for proper transcription of CBP60g, which codes for a broad regulator of immune responses that binds to the promoter of the SA biosynthesis gene ICS1. Constitutive overexpression of CBP60g restored SA accumulation and associated immunity under heat stress, demonstrating a plant with increased resilience in the face of increasing temperatures. This genotype, however, presented a late flowering phenotype, in agreement with the growth-defense trade off; the authors thus fine-tuned their engineering by linking overexpressed CBP60g with uORFsTBF1, limiting CBP60g overexpression to pathogen infection events. This uORFsTBF1–CBP60g plant demonstrated increased resilience alongside wild-type development. Interestingly, overexpression of either GBPL3 or ICS1 did not restore immunity under heat stress, the former due to heat stress specifically inhibiting GBPL3 condensate formation, and the latter likely due to the broader role that CBP60g plays in plant immune responses. Altogether, this work highlights the power of genetic engineering to develop plants that can withstand climate change, and the importance of dissecting how physiological responses operate down to a molecular level of detail. (Summary by Benjamin Jin) Nature, 10.1038/s41586-022-04902-y
Removing one wheat protein kinase provides resistance to rust fungi
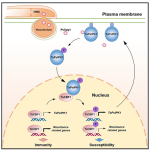
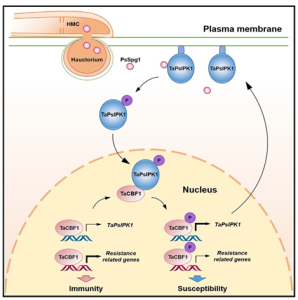 Tolerance against a pathogen can be achieved by either overexpressing a resistance-related gene, or removing a susceptibility-related gene. Overexpression of defense-related genes generally comes with side effects such as decreased growth and yield. On the other hand, a susceptibility factor might have a physiological role and its inactivation may affect agronomic traits. An effective strategy to protect plants from pathogens would be to identify and inactivate genes that promote susceptibility, but first these genes have to be found. In an effort to develop wheat that are resistant to the fungal stripe rust pathogen Puccinia striiformis f. sp. tritici (Pst), Wang et al. identified a promising susceptibility-related gene in wheat, encoding a protein kinase TaPsIPK1. They also identified a Pst effector PsSpg1 that binds to TaPsIPK1 and promotes its nuclear localization, which is crucial for the susceptibility. In the nucleus, TaPsIPK1 phosphorylates the TaCBF1d transcription factor, altering the expression of its target genes. Specifically, phosphyorylated TaCBF1d appears to both decrease expression of defense-related genes, and at the same time, enhance expression of TaPsIPK1. Overexpression of TaPsIPK1 or PsSpg1 makes wheat more susceptible to Pst. Conversely, downregulation of TaPsIPK1 in wheat provides resistance against wheat stripe rust without compromising yield, demonstrating that this susceptibility gene is a promising new tool against this fungal pathogen. Summary by Kamal Kumar Malukani, @KamalMalukani. Cell, 10.1016/j.cell.2022.06.027
Tolerance against a pathogen can be achieved by either overexpressing a resistance-related gene, or removing a susceptibility-related gene. Overexpression of defense-related genes generally comes with side effects such as decreased growth and yield. On the other hand, a susceptibility factor might have a physiological role and its inactivation may affect agronomic traits. An effective strategy to protect plants from pathogens would be to identify and inactivate genes that promote susceptibility, but first these genes have to be found. In an effort to develop wheat that are resistant to the fungal stripe rust pathogen Puccinia striiformis f. sp. tritici (Pst), Wang et al. identified a promising susceptibility-related gene in wheat, encoding a protein kinase TaPsIPK1. They also identified a Pst effector PsSpg1 that binds to TaPsIPK1 and promotes its nuclear localization, which is crucial for the susceptibility. In the nucleus, TaPsIPK1 phosphorylates the TaCBF1d transcription factor, altering the expression of its target genes. Specifically, phosphyorylated TaCBF1d appears to both decrease expression of defense-related genes, and at the same time, enhance expression of TaPsIPK1. Overexpression of TaPsIPK1 or PsSpg1 makes wheat more susceptible to Pst. Conversely, downregulation of TaPsIPK1 in wheat provides resistance against wheat stripe rust without compromising yield, demonstrating that this susceptibility gene is a promising new tool against this fungal pathogen. Summary by Kamal Kumar Malukani, @KamalMalukani. Cell, 10.1016/j.cell.2022.06.027