Plant Science Research Weekly: April 17th
Review. Beyond natural: Synthetic expansions of botanical form and function
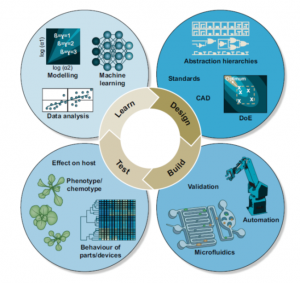 “The goal of synthetic biology is to advance the ability to dependably and consistently design or reprogram living organisms and to fabricate products from biologically-derived materials.” In this review, Patron focuses on the principles derived from engineering that are foundational to synthetic biology. She describes the central importance of standardization of all aspects, and strategies to control gene expression through synthetic inducers and light as well as synthetic gene circuits. Protein engineering can occur through directed evolution or rational design, two seemingly opposite tactics to arrive at the same outcome. Engineering metabolism in plants relies on metabolic modelling which requires an understanding of metabolite flux but is complicated by separation of reactions in different cellular compartments and even tissues. However, its compartmentalized nature can also be exploited for example to sequester toxic intermediates from synthetic pathways from the rest of the cell or concentrate enzyme substrates to increase reaction efficiency. The author provides numerous examples and discusses the potential applications of plant synthetic biology, and concludes by highlighting the need to integrate “systems biology, computational modelling and machine learning into plant biology.” (Summary by Mary Williams) New Phytol. 10.1111/nph.16562
“The goal of synthetic biology is to advance the ability to dependably and consistently design or reprogram living organisms and to fabricate products from biologically-derived materials.” In this review, Patron focuses on the principles derived from engineering that are foundational to synthetic biology. She describes the central importance of standardization of all aspects, and strategies to control gene expression through synthetic inducers and light as well as synthetic gene circuits. Protein engineering can occur through directed evolution or rational design, two seemingly opposite tactics to arrive at the same outcome. Engineering metabolism in plants relies on metabolic modelling which requires an understanding of metabolite flux but is complicated by separation of reactions in different cellular compartments and even tissues. However, its compartmentalized nature can also be exploited for example to sequester toxic intermediates from synthetic pathways from the rest of the cell or concentrate enzyme substrates to increase reaction efficiency. The author provides numerous examples and discusses the potential applications of plant synthetic biology, and concludes by highlighting the need to integrate “systems biology, computational modelling and machine learning into plant biology.” (Summary by Mary Williams) New Phytol. 10.1111/nph.16562
Review: A CRISPR way for accelerating improvement of food crops
 CRISPR technology has become indispensable for basic plant biology research and has the potential to profoundly impact future agriculture. Zhang et al. provide an overview of CRISPR technology and discuss how this technology will revolutionize future crop breeding. CRISPR technology can not only introduce DNA breaks for gene knockout, but it can also precisely edit bases. Recently, a ground-breaking genome editor “prime editing” enabled targeted insertions, deletions, and all types of point mutations, and has been successfully applied in rice and wheat. Looking forward, advanced genome editing technologies will enable plant synthetic biology, which “introduces biological components and systems that do not already exist in plants”. These can be realized in multiple ways: 1) introducing multiple edits simultaneously to “stack” beneficial traits, 2) CRISPR-mediated saturation mutagenesis to direct the evolution of new proteins and regulatory elements, and 3) accelerating crop domestication by cheap, fast, and precise gene-editing of wild crops. Finally, the authors touch upon the regulatory aspects of gene-edited crops in different countries, stressing the importance of constructing worldwide regulatory frameworks and spreading reliable information about CRISPR technology to the public to enable innovation in agriculture. (Summary by Tatsuya Nobori) Nature Food 10.1038/s43016-020-0051-8
CRISPR technology has become indispensable for basic plant biology research and has the potential to profoundly impact future agriculture. Zhang et al. provide an overview of CRISPR technology and discuss how this technology will revolutionize future crop breeding. CRISPR technology can not only introduce DNA breaks for gene knockout, but it can also precisely edit bases. Recently, a ground-breaking genome editor “prime editing” enabled targeted insertions, deletions, and all types of point mutations, and has been successfully applied in rice and wheat. Looking forward, advanced genome editing technologies will enable plant synthetic biology, which “introduces biological components and systems that do not already exist in plants”. These can be realized in multiple ways: 1) introducing multiple edits simultaneously to “stack” beneficial traits, 2) CRISPR-mediated saturation mutagenesis to direct the evolution of new proteins and regulatory elements, and 3) accelerating crop domestication by cheap, fast, and precise gene-editing of wild crops. Finally, the authors touch upon the regulatory aspects of gene-edited crops in different countries, stressing the importance of constructing worldwide regulatory frameworks and spreading reliable information about CRISPR technology to the public to enable innovation in agriculture. (Summary by Tatsuya Nobori) Nature Food 10.1038/s43016-020-0051-8
Review: The domestication syndrome in vegetatively propagated field crops
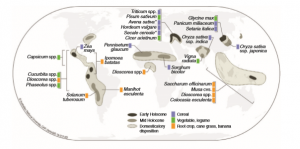 Archeological remains and genetic analyses of ancient DNA have revealed that the loss of seed dispersal marks the onset of domestication for sexually reproduced crops such as cereal, legume and oils seed crops, for which annual cultivation is based on sowing seeds. In contrast, less is known about the domestication of vegetatively propagating plants including sugarcane, bananas and root crops, for which perennial cultivation relies on clones obtained by parthenogenesis or regeneration from fragments of vegetative structures. In Denham et al., the authors pinpoint the shift towards asexual reproduction as the main convergent evolutionary trait in vegetatively propagated crops; humans selected variants with reproductive dysfunction that originated through natural processes (e.g., spontaneous mutations, hybridization and polyploidization), and propagated plants with seedless fruits and sterile polyploid (including triploid banana and polyploid sugarcane). Other traits associated with domestication include increased yield of the edible portion and greater ease of harvesting. Vegetatively propagated plants show great phenotypic variation which can be associated with developmental plasticity rather than genotypic diversity, therefore, many morphological changes may be due to dynamic expression of phenotypic traits in response to growth environment and cultivation practices. (Summary by Michela Osnato) Ann. Bot. 10.1093/aob/mcz212
Archeological remains and genetic analyses of ancient DNA have revealed that the loss of seed dispersal marks the onset of domestication for sexually reproduced crops such as cereal, legume and oils seed crops, for which annual cultivation is based on sowing seeds. In contrast, less is known about the domestication of vegetatively propagating plants including sugarcane, bananas and root crops, for which perennial cultivation relies on clones obtained by parthenogenesis or regeneration from fragments of vegetative structures. In Denham et al., the authors pinpoint the shift towards asexual reproduction as the main convergent evolutionary trait in vegetatively propagated crops; humans selected variants with reproductive dysfunction that originated through natural processes (e.g., spontaneous mutations, hybridization and polyploidization), and propagated plants with seedless fruits and sterile polyploid (including triploid banana and polyploid sugarcane). Other traits associated with domestication include increased yield of the edible portion and greater ease of harvesting. Vegetatively propagated plants show great phenotypic variation which can be associated with developmental plasticity rather than genotypic diversity, therefore, many morphological changes may be due to dynamic expression of phenotypic traits in response to growth environment and cultivation practices. (Summary by Michela Osnato) Ann. Bot. 10.1093/aob/mcz212
Review: Delayed luminescence of seeds: are shining seeds viable?
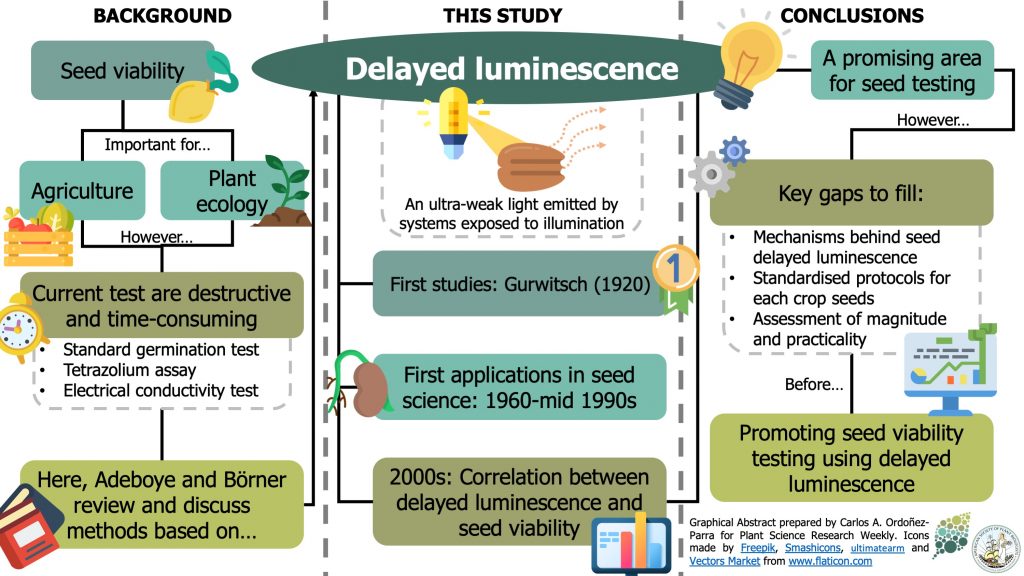 Most of the current methods for assessing seed viability are time-consuming and destructive. As an alternative, some authors have suggested using procedures based on delayed luminescence; an ultra-weak light emitted by biological materials exposed to illumination. Here, Adeboye and Börner review the history of seed viability testing, the milestones in the study of delayed luminescence, and future directions to start using it in seed science. The authors track the study of delayed luminescence to 1920 and its first applications in seed science between 1960 and the mid-1990s. Then, they show how, since the 2000s, some studies found significant correlations between different aspects of delayed luminescence (i.e., intensity, spectra, and time) and seed viability. The authors argue that the study of delayed luminescence in seeds is a promising area and outline some gaps that need to be addressed before validating this technique for seed testing. (Summary by Carlos A. Ordóñez-Parra) Seed Sci. Technol. 10.15258/sst.2020.48.2.04
Most of the current methods for assessing seed viability are time-consuming and destructive. As an alternative, some authors have suggested using procedures based on delayed luminescence; an ultra-weak light emitted by biological materials exposed to illumination. Here, Adeboye and Börner review the history of seed viability testing, the milestones in the study of delayed luminescence, and future directions to start using it in seed science. The authors track the study of delayed luminescence to 1920 and its first applications in seed science between 1960 and the mid-1990s. Then, they show how, since the 2000s, some studies found significant correlations between different aspects of delayed luminescence (i.e., intensity, spectra, and time) and seed viability. The authors argue that the study of delayed luminescence in seeds is a promising area and outline some gaps that need to be addressed before validating this technique for seed testing. (Summary by Carlos A. Ordóñez-Parra) Seed Sci. Technol. 10.15258/sst.2020.48.2.04
How to transfer lipids from one membrane to another during thylakoid biogenesis
 The thylakoid membranes are located in the stroma of chloroplasts and house the machinery for the photosynthetic light reactions. They emerge largely de novo during the transition from pro-plastids into mature, photosynthesizing chloroplasts. Generating new thylakoid membranes requires a supply of lipids, the biosynthesis of which is thought to occur on the chloroplast inner envelope. Different models, involving direct membrane contact sites, vesicle trafficking, and lipid-binding proteins, have been proposed to explain how the lipids are transferred from the envelope to the thylakoids during periods of thylakoid membrane expansion. By searching the Arabidopsis genome for candidates, Hertle et al. identified a predicted lipid-binding protein (CPSFL1), which localizes to the chloroplast and is essential for photoautotrophic growth. Electron microscopic imaging showed a severely underdeveloped thylakoid network in cpsfl1 mutant plants. The mutant chloroplasts also seemed to completely lack vesicles normally seen at/near the inner envelope in the wild type. Combined with data from in vitro lipid binding assays, these results indicate that CPSFL1 acts in a chloroplast vesicular trafficking pathway that plays a major role in thylakoid membrane biogenesis. (Summary by Frej Tulin) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1916946117
The thylakoid membranes are located in the stroma of chloroplasts and house the machinery for the photosynthetic light reactions. They emerge largely de novo during the transition from pro-plastids into mature, photosynthesizing chloroplasts. Generating new thylakoid membranes requires a supply of lipids, the biosynthesis of which is thought to occur on the chloroplast inner envelope. Different models, involving direct membrane contact sites, vesicle trafficking, and lipid-binding proteins, have been proposed to explain how the lipids are transferred from the envelope to the thylakoids during periods of thylakoid membrane expansion. By searching the Arabidopsis genome for candidates, Hertle et al. identified a predicted lipid-binding protein (CPSFL1), which localizes to the chloroplast and is essential for photoautotrophic growth. Electron microscopic imaging showed a severely underdeveloped thylakoid network in cpsfl1 mutant plants. The mutant chloroplasts also seemed to completely lack vesicles normally seen at/near the inner envelope in the wild type. Combined with data from in vitro lipid binding assays, these results indicate that CPSFL1 acts in a chloroplast vesicular trafficking pathway that plays a major role in thylakoid membrane biogenesis. (Summary by Frej Tulin) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1916946117
miRNA-mediated lateral inhibition controls rhizoid cell patterning in Marchantia polymorpha
 In multicellular organisms, the patterning of different cell types in spatial arrays is regulated through several mechanisms, one of which is lateral inhibition, a process well characterized in metazoans. In this process, an individual cell transmits signals to neighboring cells to instruct a different cell fate to that of the instructing cell. In plants, lateral inhibition mechanisms described to-date (e.g., guard cell and trichome patterning) act mainly through mobile proteins. In a recent paper Thamm et al., describe an activator-repressor system that regulates rhizoid cell patterning through lateral inhibition in Marchantia polymorpha. The authors make use of simple cellular automata models and genetic data to describe a system where the FEW RHIZOIDS (MpFRH1) microRNA regulates cell fate in neighboring rhizoid cells through repression of the ROOT HAIR DEFECTIVE SIX-LIKE1 (MpRSL1) transcription factor, a positive regulator of MpFRH1 essential for rhizoid development. The authors demonstrate that, in contrast to wild-type plants where rhizoid cells form linear chain-like structures, Mpfrh1loss of function mutants develop two-dimensional clusters of rhizoid cells. The authors propose a model in which epidermal cells are inhibited from adopting the rhizoid cell fate only when these are adjacent to two rhizoid cells, and so exposed to more than a threshold level of MpFRH1 repressor. The authors observe that “the MpFRH1-lateral inhibition mechanism is restricted to the liverworts, suggesting that different lateral inhibition mechanisms control the patterning of rhizoids in liverworts, and root hairs in angiosperms.” (Summary by Jesus Leon) Curr. Biol. 10.1016/j.cub.2020.03.032
In multicellular organisms, the patterning of different cell types in spatial arrays is regulated through several mechanisms, one of which is lateral inhibition, a process well characterized in metazoans. In this process, an individual cell transmits signals to neighboring cells to instruct a different cell fate to that of the instructing cell. In plants, lateral inhibition mechanisms described to-date (e.g., guard cell and trichome patterning) act mainly through mobile proteins. In a recent paper Thamm et al., describe an activator-repressor system that regulates rhizoid cell patterning through lateral inhibition in Marchantia polymorpha. The authors make use of simple cellular automata models and genetic data to describe a system where the FEW RHIZOIDS (MpFRH1) microRNA regulates cell fate in neighboring rhizoid cells through repression of the ROOT HAIR DEFECTIVE SIX-LIKE1 (MpRSL1) transcription factor, a positive regulator of MpFRH1 essential for rhizoid development. The authors demonstrate that, in contrast to wild-type plants where rhizoid cells form linear chain-like structures, Mpfrh1loss of function mutants develop two-dimensional clusters of rhizoid cells. The authors propose a model in which epidermal cells are inhibited from adopting the rhizoid cell fate only when these are adjacent to two rhizoid cells, and so exposed to more than a threshold level of MpFRH1 repressor. The authors observe that “the MpFRH1-lateral inhibition mechanism is restricted to the liverworts, suggesting that different lateral inhibition mechanisms control the patterning of rhizoids in liverworts, and root hairs in angiosperms.” (Summary by Jesus Leon) Curr. Biol. 10.1016/j.cub.2020.03.032
AT-hook transcription factors restrict petiole elongation by inhibiting PIF-activated genes
 The AT-hook motif nuclear localized (AHL) transcription factor family has one or two DNA-binding motifs to bind to AT rich DNA regions, and they also have a conserved PPC-DUF domain for protein-protein interactions. AHLs affect a wide range of biotic and abiotic responses but the mechanism of how they regulate transcription are still not well understood. Favero et al. report the first ChIP-seq analysis of an AHL family member, SOB3/AHL29, and show that AHL29 represses the transcription of PIF-regulated (Phytochrome-interacting factor) genes. Consistent with this finding, the sob3-6 mutant has increased petiole length while the pif4 mutant has the opposite phenotype, and the transcriptomes of AHL and PIF mutants show that AHL oppositely affects the transcription of large numbers PIF-regulated genes. Furthermore, ChIP-seq of AHL29 and PIF4/5 reveal that both proteins bind to similar regions in their target genes. Taken together, the authors conclude that AHLs repress petiole growth by decreasing PIF occupancy at growth-promoting genes through inhibiting the transcriptional activation. (Summary by Min May Wong) Curr. Biol. 10.1016/j.cub.2020.02.017
The AT-hook motif nuclear localized (AHL) transcription factor family has one or two DNA-binding motifs to bind to AT rich DNA regions, and they also have a conserved PPC-DUF domain for protein-protein interactions. AHLs affect a wide range of biotic and abiotic responses but the mechanism of how they regulate transcription are still not well understood. Favero et al. report the first ChIP-seq analysis of an AHL family member, SOB3/AHL29, and show that AHL29 represses the transcription of PIF-regulated (Phytochrome-interacting factor) genes. Consistent with this finding, the sob3-6 mutant has increased petiole length while the pif4 mutant has the opposite phenotype, and the transcriptomes of AHL and PIF mutants show that AHL oppositely affects the transcription of large numbers PIF-regulated genes. Furthermore, ChIP-seq of AHL29 and PIF4/5 reveal that both proteins bind to similar regions in their target genes. Taken together, the authors conclude that AHLs repress petiole growth by decreasing PIF occupancy at growth-promoting genes through inhibiting the transcriptional activation. (Summary by Min May Wong) Curr. Biol. 10.1016/j.cub.2020.02.017
Self-activation loop of STM maintains the meristematic activity of cells at leaf axil
 Plasticity in plant development comes from the meristematic cells that are maintained throughout plant growth and among other things produce lateral organs such as shoot branches. In this paper, Cao et al. show that the self-activation loop of SHOOT MERISTEMLESS (STM), a meristem marker gene that encodes a homeodomain transcription factor, is important for the maintenance of meristematic cells at the leaf axil. The authors identified a mutant, ath1-3, with a T-DNA insertion in ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) gene, encoding a BEL1 like homeodomain protein, that fails to initiate axillary meristems. ATH1 is expressed broadly in the shoot apical meristem and in leaf primordia. In the ath1-3 mutant, the expression of STM decreased in the axils of leaves produced later in development compared to Col-0. Both ATH1 and STM are nuclear localized and bind to the overlapping cis elements in the STM locus, activating its expression. Increased expression of STM was observed only when both ATH1 and STM were present compared to either of them alone, suggesting the formation of heterodimers in self-activation loop. Competition experiments with the KNOX domain of STM, which retains the interaction with ATH1 but has reduced transcriptional activity, confirmed the role of ATH1-STM heterodimer in STM activation. Basal expression of STM in the leaf axil cells maintains the chromatin in permissive state; Histone3 lysine 27 trimethylation (H3K27me3), a signature of silenced chromatin, was increased, and acetylation marks on Histone3 (H3Ac), signatures of active chromatin, were decreased in ath1-3 compared to Col-0. (Summary by Vijaya Batthula) Curr. Biol. 10.1016/j.cub.2020.03.031
Plasticity in plant development comes from the meristematic cells that are maintained throughout plant growth and among other things produce lateral organs such as shoot branches. In this paper, Cao et al. show that the self-activation loop of SHOOT MERISTEMLESS (STM), a meristem marker gene that encodes a homeodomain transcription factor, is important for the maintenance of meristematic cells at the leaf axil. The authors identified a mutant, ath1-3, with a T-DNA insertion in ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) gene, encoding a BEL1 like homeodomain protein, that fails to initiate axillary meristems. ATH1 is expressed broadly in the shoot apical meristem and in leaf primordia. In the ath1-3 mutant, the expression of STM decreased in the axils of leaves produced later in development compared to Col-0. Both ATH1 and STM are nuclear localized and bind to the overlapping cis elements in the STM locus, activating its expression. Increased expression of STM was observed only when both ATH1 and STM were present compared to either of them alone, suggesting the formation of heterodimers in self-activation loop. Competition experiments with the KNOX domain of STM, which retains the interaction with ATH1 but has reduced transcriptional activity, confirmed the role of ATH1-STM heterodimer in STM activation. Basal expression of STM in the leaf axil cells maintains the chromatin in permissive state; Histone3 lysine 27 trimethylation (H3K27me3), a signature of silenced chromatin, was increased, and acetylation marks on Histone3 (H3Ac), signatures of active chromatin, were decreased in ath1-3 compared to Col-0. (Summary by Vijaya Batthula) Curr. Biol. 10.1016/j.cub.2020.03.031
Self-isolation to the rescue – how plasmodesmata initiate signaling in response to chitin
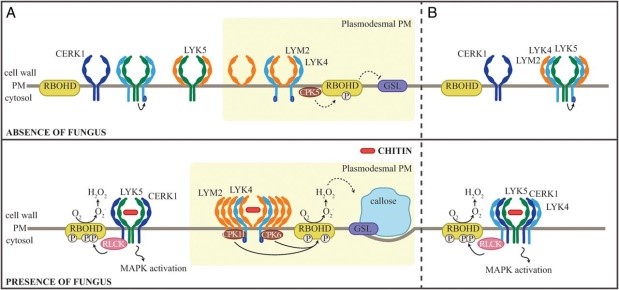 Plant cells are known to initiate local and systemic signaling in response to certain stressful conditions to safeguard cells in the immediate vicinity as well as that are far from the site of stimuli. In this study, Cheval and co-workers have characterized a pathway in plant cells for sensing and responding to chitin, a polysaccharide and chief constituent of fungal cell walls. This pathway occurs in plasmodesmata, the tunnels that connect adjacent cells through symplastic transport systems. In response to chitin, a receptor-kinase pair of LYM2-LYK4 has been shown to get activated, enabling callose deposition to plug plasmodematal connections, resulting in the isolation of individual cells. Curiously, this callose deposition also depends on two calcium-dependent protein kinases (CDPKs) and the NADPH oxidase, RBOHD, though connections between LYM2-LYK4 and these proteins are yet to be deciphered. These results imply the need for Ca2+ and/or ROS signals, if any, to enable plugging of plasmodemata, which needs further experimental validation. The authors summarize, “This work characterizes how a cell can produce a localized and specific response in a discrete membrane domain, identifying that there is microdomain specificity in immune signaling to a single elicitor and that cell-to-cell connections are independently controlled. “ (Summary by Pavithran Narayanan) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1907799117
Plant cells are known to initiate local and systemic signaling in response to certain stressful conditions to safeguard cells in the immediate vicinity as well as that are far from the site of stimuli. In this study, Cheval and co-workers have characterized a pathway in plant cells for sensing and responding to chitin, a polysaccharide and chief constituent of fungal cell walls. This pathway occurs in plasmodesmata, the tunnels that connect adjacent cells through symplastic transport systems. In response to chitin, a receptor-kinase pair of LYM2-LYK4 has been shown to get activated, enabling callose deposition to plug plasmodematal connections, resulting in the isolation of individual cells. Curiously, this callose deposition also depends on two calcium-dependent protein kinases (CDPKs) and the NADPH oxidase, RBOHD, though connections between LYM2-LYK4 and these proteins are yet to be deciphered. These results imply the need for Ca2+ and/or ROS signals, if any, to enable plugging of plasmodemata, which needs further experimental validation. The authors summarize, “This work characterizes how a cell can produce a localized and specific response in a discrete membrane domain, identifying that there is microdomain specificity in immune signaling to a single elicitor and that cell-to-cell connections are independently controlled. “ (Summary by Pavithran Narayanan) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1907799117
Subtilase activity in intrusive cells mediates haustorium maturation in parasitic plants
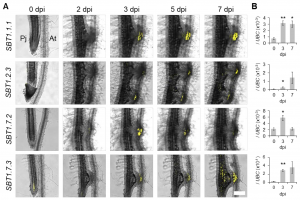 Parasitic plants develop unique structures called haustoria that penetrate into the host plant vasculature, from which they take nutrients. During this process, haustorial epidermal cells differentiate into specialized cells called intrusive cells, which eventually re-differentiate into a xylem bridge connected to the host vasculature. In Ogawa et al., the authors identified differentially expressed genes in the intrusive cells of the model parasitic plant, Phtheirospermum japonicum. RNA-seq data of laser-dissected haustoria revealed genes highly upregulated in intrusive cells as shown by promoter expression, allowing the authors to obtain marker genes specifically expressed in intrusive cells. Additionally, the authors identified four genes encoding subtilisin-like serine proteases (SBTs) that are highly expressed in intrusive cells. Expressing the SBT inhibitor Epi10 (Extracellular proteinase inhibitor 10) in the intrusive cells interfered with xylem bridge formation as well as auxin signaling. These findings indicate that subtilase genes expressed in intrusive cells are required for the auxin-dependent maturation of haustoria during host-parasite interaction. (Summary by Sunita Pathak) bioRxiv: 10.1101/2020.03.30.015149
Parasitic plants develop unique structures called haustoria that penetrate into the host plant vasculature, from which they take nutrients. During this process, haustorial epidermal cells differentiate into specialized cells called intrusive cells, which eventually re-differentiate into a xylem bridge connected to the host vasculature. In Ogawa et al., the authors identified differentially expressed genes in the intrusive cells of the model parasitic plant, Phtheirospermum japonicum. RNA-seq data of laser-dissected haustoria revealed genes highly upregulated in intrusive cells as shown by promoter expression, allowing the authors to obtain marker genes specifically expressed in intrusive cells. Additionally, the authors identified four genes encoding subtilisin-like serine proteases (SBTs) that are highly expressed in intrusive cells. Expressing the SBT inhibitor Epi10 (Extracellular proteinase inhibitor 10) in the intrusive cells interfered with xylem bridge formation as well as auxin signaling. These findings indicate that subtilase genes expressed in intrusive cells are required for the auxin-dependent maturation of haustoria during host-parasite interaction. (Summary by Sunita Pathak) bioRxiv: 10.1101/2020.03.30.015149
DNA methylation and integrity in aged seeds and regenerated plants ($)
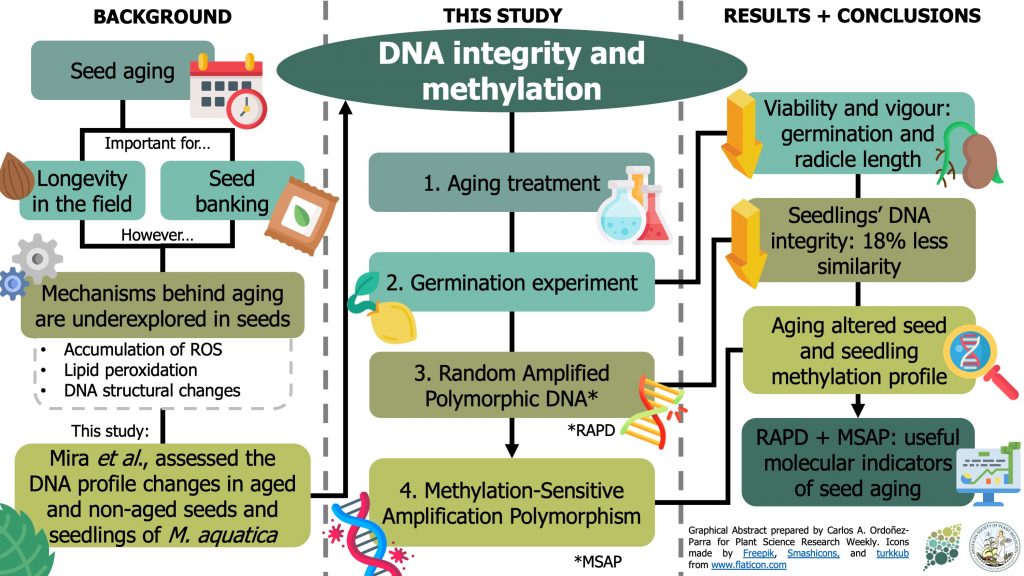 Understanding seed aging is crucial to comprehend seed longevity in the field and to design ex situ conservation programs. However, the mechanisms behind seed aging are underexplored. In this paper, Mira et al. assessed the changes in DNA integrity and methylation between aged and non-aged seeds of Mentha aquatica and the seedlings obtained from these seeds. To achieve these, they used Random Amplified Polymorphic DNA (RAPD) and Methylation-Sensitive Amplification Polymorphism (MSAP) analysis to asses changes in the DNA profile of the seeds and seedlings after the aging treatment. Surprisingly, DNA integrity was only affected in the seedlings from aged seeds, suggesting the loss of DNA integrity might be undetectable in the seed stage. However, aged seed and seedlings had an altered methylation profiles. These results indicate that seed aging in M. aquatica is associated with changes in DNA methylation in seeds and seedlings, and the accumulation of DNA lesions in seedlings. Also, the authors suggest the use of RAPD and MSAP as molecular indicators to continue studying seed aging (Summary by Carlos A. Ordóñez-Parra) Seed Sci. Res. 10.1017/S0960258520000021
Understanding seed aging is crucial to comprehend seed longevity in the field and to design ex situ conservation programs. However, the mechanisms behind seed aging are underexplored. In this paper, Mira et al. assessed the changes in DNA integrity and methylation between aged and non-aged seeds of Mentha aquatica and the seedlings obtained from these seeds. To achieve these, they used Random Amplified Polymorphic DNA (RAPD) and Methylation-Sensitive Amplification Polymorphism (MSAP) analysis to asses changes in the DNA profile of the seeds and seedlings after the aging treatment. Surprisingly, DNA integrity was only affected in the seedlings from aged seeds, suggesting the loss of DNA integrity might be undetectable in the seed stage. However, aged seed and seedlings had an altered methylation profiles. These results indicate that seed aging in M. aquatica is associated with changes in DNA methylation in seeds and seedlings, and the accumulation of DNA lesions in seedlings. Also, the authors suggest the use of RAPD and MSAP as molecular indicators to continue studying seed aging (Summary by Carlos A. Ordóñez-Parra) Seed Sci. Res. 10.1017/S0960258520000021
Early Holocene crop cultivation and landscape modification in Amazonia
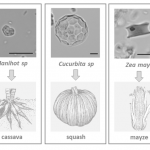
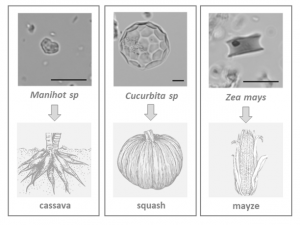 To date, archeobotanical and genetic studies indicated four centers of plant cultivation in the early Holocene: China and the Near East in the Old World, and southwestern Mexico and northwestern South America in the New World. Recent research of Lombardo et al. revealed that Llanos de Moxos (Bolivia) in southwestern Amazonia could also be a hotspot of plant domestication. From 10,850 calibrated years before present (cal. yr bp), the first inhabitants of this region created artificial forest islands within seasonally flooded savannah. As revealed by the analysis of microfossils found in surveyed sites, these fertile patches (with an average size of 0.5 hectares) were probably the ecosystem in which human settlers domesticated important plant species including manioc (cassava), squash and maize. From this region, the oldest evidence of plant cultivation came from the identification of: heart-shaped phytolith related to Manihot sp. dated to 10,350 cal. yr bp, spherical phytoliths derived from the rind of Cucurbita sp. in layers dated to 10,250 cal yr bp, and rondel phytoliths produced in the cob of a partially domesticated variety of maize dated to 6,850 cal. yr bp. The authors outline how humans have engineered the environment for farming since they colonized Amazonia in the early Holocene. Nowadays, these human-made forest islands play crucial roles in the conservation of biodiversity, as they serve as feeding and roosting sites for some threatened bird species. (Summary by Michela Osnato) Nature 10.1038/s41586-020-2162-7
To date, archeobotanical and genetic studies indicated four centers of plant cultivation in the early Holocene: China and the Near East in the Old World, and southwestern Mexico and northwestern South America in the New World. Recent research of Lombardo et al. revealed that Llanos de Moxos (Bolivia) in southwestern Amazonia could also be a hotspot of plant domestication. From 10,850 calibrated years before present (cal. yr bp), the first inhabitants of this region created artificial forest islands within seasonally flooded savannah. As revealed by the analysis of microfossils found in surveyed sites, these fertile patches (with an average size of 0.5 hectares) were probably the ecosystem in which human settlers domesticated important plant species including manioc (cassava), squash and maize. From this region, the oldest evidence of plant cultivation came from the identification of: heart-shaped phytolith related to Manihot sp. dated to 10,350 cal. yr bp, spherical phytoliths derived from the rind of Cucurbita sp. in layers dated to 10,250 cal yr bp, and rondel phytoliths produced in the cob of a partially domesticated variety of maize dated to 6,850 cal. yr bp. The authors outline how humans have engineered the environment for farming since they colonized Amazonia in the early Holocene. Nowadays, these human-made forest islands play crucial roles in the conservation of biodiversity, as they serve as feeding and roosting sites for some threatened bird species. (Summary by Michela Osnato) Nature 10.1038/s41586-020-2162-7