Recognizing featured Plant Cell first authors, February 2017
Masanori Izumi, featured first author of Entire Photodamaged Chloroplasts Are Transported to the Central Vacuole by Autophagy
 Current Position: Assistant Professor, Frontier Research Institute for Interdisciplinary Sciences, Tohoku University.
Current Position: Assistant Professor, Frontier Research Institute for Interdisciplinary Sciences, Tohoku University.
Education: Ph.D. (2012), Graduate School of Agricultural Sciences, Tohoku University, Japan.
Non-scientific Interests: Playing tennis, Travel to Japanese hot springs.
When I was an undergraduate student, I joined Dr. Hiroyuki Ishida’s research group, which is working to identify the degradation mechanism of chloroplast proteins during leaf senescence. This gave me a chance to realize the interesting nature of chloroplasts. I performed research demonstrating that chloroplasts are partially degraded via a type of autophagic vesicles known as Rubisco-containing bodies (RCBs). This included two collaborative works with Prof. Yoshinori Ohsumi, a winner of the 2016 Nobel Prize in Physiology or Medicine. In my Ph.D. research, I revealed the physiological importance of RCB pathway under energy-limited conditions. As a post-doc, I was awarded a grant for young scientists from the Japanese government as a hosted researcher in Dr. Jun Hidema’s Lab, in which the members are focused on plant response to ultraviolet-B (UVB) damage. I exposed Arabidopsis autophagy-deficient mutants to UVB and found that they showed a UVB-sensitive phenotype. Because I then expected that the RCB pathway was specifically activated by UVB damage, I carried out detailed imaging analysis with confocal microscopy. Although this original expectation was not true, I noticed that chlorophagy, the vacuolar transport and destruction of entire chloroplasts by autophagy, occurs in UVB-damaged leaves. This is the topic of our current article. During this work, I became an assistant professor as an independent researcher. Our small group still has many questions about chlorophagy such as how photodamaged chloroplasts are recognized and recruited for autophagy. We are attempting to elucidate these and other interesting questions.
Qiong Nan and Dong Qian, featured first authors of Plant Actin-depolymerizing Factors Possess Opposing Biochemical Properties Arising From Key Amino Acid Changes Throughout Evolution.
 Qiong Nan
Qiong Nan
Current Position: Ph.D. student at the School of Life Sciences, Lanzhou University, China.
Education: B.S. (2011) in Biology, College of Bioengineering and Biotechnology, Tianshui Normal University, China.
Non-scientific Interests: Reading, playing football and listening to music.
I spent most of my childhood on farms, where I had ample opportunity to learn about various plants. While growing up, the wide variety of plants present in farms and other places created enough curiosity in my mind to lead me to choose Biology as my major at Tianshui Normal University. After earning my B.S., I had the opportunity to join Dr. Xiang’s lab at Lanzhou University as a graduate student. I was accepted as a Ph.D. candidate under the guidance of Dr. Yun Xiang and Dr. Lizhe An, once I finished my Master’s course work. My research has focused on investigating the functions of Actin-depolymerizing factors (ADFs) and other actin binding proteins in plants. When I joined the lab, Jingen Zhu, a graduate student in the lab, had found that the F-actin bundling activity of AtADF5 differed from that of other conserved ADFs. I was fortunate to be given the opportunity to investigate the functional evolution of ADF family members across various plant lineages. As reported in this paper, the evolution of these N-terminal extensions, arising from intron sliding events and several conserved mutations, have produced the diverse biochemical functions of plant ADFs from a putative ancestor. I hope the key findings of this project contribute to our understanding of ADFs and their co-evolution with actin.
Dong Qian
 Current Position: Postdoctoral Researcher, School of Life Sciences, Lanzhou University, China.
Current Position: Postdoctoral Researcher, School of Life Sciences, Lanzhou University, China.
Education: Ph.D. (2016) in Plant Biology and B.S. (2009) in Biology, School of Life Sciences, Lanzhou University, China.
Non-scientific Interests: Reading, running and playing table tennis.
During my studies at Lanzhou University, I was fascinated to learn how plants make developmental decisions by perceiving and responding to environmental signaling. I was fortunate to begin my studies in plant science as an M.S. student under the guidance of Dr. Yun Xiang. After two years of my master course, I was honored to work with Dr. Yun Xiang and Dr. Lizhe An as a Ph.D. candidate. During my research, I worked on the molecular mechanisms underlying remodeling of the actin cytoskeleton and the mechanisms of plant responses to abiotic stress through regulating and remodeling of the actin cytoskeleton. In due course, I developed a solid foundation in biological research through theoretical and practical training. At present, I am working as a postdoctoral researcher under the supervision of Dr. Yun Xiang. Here, we investigate the mechanisms of functional divergence among Actin-depolymerizing factors (ADFs) and identify key sites associated with their diverse biochemical functions. We hope this work provides new insights into the molecular mechanisms underlying the evolution of ADF family members across plant lineages and will be valuable for researchers working on ADFs.
Yan Zhu, Liang Rong and Qiang Luo, featured first authors of The Histone Chaperone NRP1 Interacts With WEREWOLF to Activate GLABRA2 in Arabidopsis Root Hair Development
 Yan Zhu
Yan Zhu
Current Position: Associate Professor in State Key Laboratory of Genetic Engineering, Department of Biochemistry, Institute of Plant Biology, School of Life Sciences, Fudan University, Shanghai, China.
Education: Ph.D. (2006) in IBMP-CNRS, France.
Non-scientific Interests: Walking and table tennis.
My academic research started from the cDNA cloning of one rice histone chaperone, OsNAP1, which is a conserved factor binding histones and facilitating their incorporation into nucleosomes. Since then, I have become interested in and focused on the machineries and mechanisms involved in the dynamic nucleosome assembly and disassembly in plants. I work on plant histone chaperones and chromatin remodeling factors, and characterize their roles in plant growth and development, in genome stability, and in the interplay with the environment. By using a wide range of techniques, we found that histone chaperone NRP1 specifically interacts with the transcription factor WER, mediates the downstream activation of GL2, and further affects cell fate determination in the root epidermis.
 Liang Rong
Liang Rong
Current Position: Postdoctoral Research Associate, Dept. of Pharmacology & Pharmaceutical Sciences, School of Pharmacy, University of Southern California.
Education: Ph.D. (2015) in Biochemistry and Molecular biology, School of Life Sciences, Fudan University, Shanghai, P. R. China.
Non-scientific Interests: Basketball, table tennis, travelling, literature, and music.
After I got my bachelor’s degree at University of Science and Technology in Beijing, I changed my major from mineral resources engineering to life sciences. I pursued my interest in biochemistry and molecular biology and got my master’s degree at Anhui Agricultural University in 2011. Then I continued to my Ph.D. study in Dr. Jinbiao Ma’s lab at Fudan University and my research area is X-ray crystallography. I collaborated with Dr. Yan Zhu in Dr. Aiwu Dong’s lab to examine AtNRP1 function in root hair development. I was attracted by the idea that a histone chaperone can interact with a transcription factor to promote gene expression. First, I expressed recombinant AtH2A/H2B, AtH3/H4, AtNRP1, and WER in E. coli. Then I resolved the crystal structure of AtNPR1, conducted biochemical assays, and tried to propose a model to explain the mechanism how AtNRP1/NRP2 regulate GL2 expression. This project gave me the opportunity to learn many techniques in biochemistry, molecular biology and structural biology. I started my postdoc research in school of pharmacy at the University of Southern California in 2015.
 Qiang Luo
Qiang Luo
Current Position: Ph.D. student in State Key Laboratory of Genetic Engineering, Department of Biochemistry, Institute of Plant Biology, School of Life Sciences, Fudan University, Shanghai, China.
Education: B.S (2014) in Biology Engineering at Lanzhou University, Lanzhou, China.
Non-scientific Interests: Camping, playing table tennis, and watching movies.
I was born in a small village in Sichun province of China. When I was in my high school, I found that biology is full of mysteries, which is really attractive and amazing to me. So I chose biology as my major in Lanzhou University. Later I continued my biological research at Fudan University in the labs of Dr. Aiwu Dong, Dr. Wen-Hui Shen, and Dr. Jinbiao Ma. We are very interested in molecular mechanisms of epigenetic regulation, especially the function of histone chaperones and histone modifications. I mainly worked on biochemical experiments to explore the interactions among the histone chaperone NRP1, the transcription factor WER, and the GL2 promoter. I will continue to explore the complex structures of histone chaperones with histones and with transcription factors in the future.


 During vegetative growth, the shoot apical meristem (SAM) produces lateral organ primordia but remains roughly the same size, as WUSCHEL–CLAVATA signaling modulates the balance between cell division and differentiation. During the transition to reproductive growth in many species, the SAM expands in size and becomes domed in shape, as if puffing itself up in anticipation of the crucial job of producing flowers (reviewed in
During vegetative growth, the shoot apical meristem (SAM) produces lateral organ primordia but remains roughly the same size, as WUSCHEL–CLAVATA signaling modulates the balance between cell division and differentiation. During the transition to reproductive growth in many species, the SAM expands in size and becomes domed in shape, as if puffing itself up in anticipation of the crucial job of producing flowers (reviewed in Crowdsourcing leverages the inputs, ideas and talents of many people in parallel to achieve a goal. Crowdsourcing or “citizen science” can also be an effective approach for science outreach that engages the public by enabling them to collect and analyze data. Willis et al. have developed and assessed a crowdsourced project in which digitized herbaria specimens were evaluated to generate a phenology database. Specifically, specimens of two species collected over 200 years were analyzed for the numbers of flower buds, flowers and fruits, to investigate changes in the timing of flowering. The authors found no differences in the quality of the data collected by experts and non-experts, demonstrating the feasibility of this approach. The project lives on the portal
Crowdsourcing leverages the inputs, ideas and talents of many people in parallel to achieve a goal. Crowdsourcing or “citizen science” can also be an effective approach for science outreach that engages the public by enabling them to collect and analyze data. Willis et al. have developed and assessed a crowdsourced project in which digitized herbaria specimens were evaluated to generate a phenology database. Specifically, specimens of two species collected over 200 years were analyzed for the numbers of flower buds, flowers and fruits, to investigate changes in the timing of flowering. The authors found no differences in the quality of the data collected by experts and non-experts, demonstrating the feasibility of this approach. The project lives on the portal 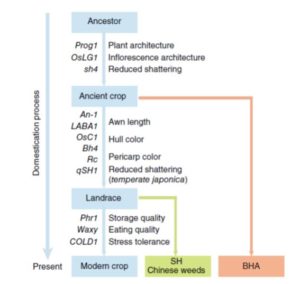 Crop domestication has been accompanied by the evolution of aggressive weedy crop relatives that compete for resources and make weed management a challenge. By using whole-genome sequencing of the two most commonly found weedy rice strains in the US (SH and BHA), and comparing them with the genomes of several accessions of cultivated rice, wild progenitors and other weedy strains from China, Li et al. aimed to shed light on the genetic mechanisms underlying the evolution of weedy rice. The authors found that US weedy rice strains originated from Asian accessions after the domestication of rice in Asia, finding support for the de-domestication hypothesis due to the presence of certain early-selected domestication-associated alleles. Moreover, they found that weediness can apparently evolve relatively easily, through selection on a small number of genomic regions. These findings should be a warning to rice producers around the world to stay vigilant to the presence of weedy strains. (Summary by
Crop domestication has been accompanied by the evolution of aggressive weedy crop relatives that compete for resources and make weed management a challenge. By using whole-genome sequencing of the two most commonly found weedy rice strains in the US (SH and BHA), and comparing them with the genomes of several accessions of cultivated rice, wild progenitors and other weedy strains from China, Li et al. aimed to shed light on the genetic mechanisms underlying the evolution of weedy rice. The authors found that US weedy rice strains originated from Asian accessions after the domestication of rice in Asia, finding support for the de-domestication hypothesis due to the presence of certain early-selected domestication-associated alleles. Moreover, they found that weediness can apparently evolve relatively easily, through selection on a small number of genomic regions. These findings should be a warning to rice producers around the world to stay vigilant to the presence of weedy strains. (Summary by 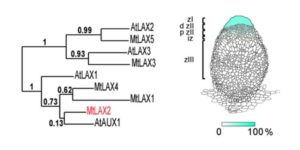 Nodules are specialized structures that form symbiotic, nitrogen-fixing associations with rhizobia on some plants including Medicago truncatula. Previous work has shown that auxin signalling is involved in nodule formation. Roy et al. extend this knowledge through the identification of a Medicago gene encoding MtLAX2 that is a functional homologue of the Arabidopsis auxin influx transporter AUX1. Loss of function mutants mtlax2 and aux1 show similar phenotypes including reduced sensitivity to auxin, reduced lateral root number, and reduced root gravitropism. Furthermore, MtLAX2 is expressed at very early stages of nodule formation and is required for nodule initiation. This study indicates that auxin influx is crucial for both nodule and lateral root organogenesis, and provides additional insights into the process of nodule formation. Plant Physiol.
Nodules are specialized structures that form symbiotic, nitrogen-fixing associations with rhizobia on some plants including Medicago truncatula. Previous work has shown that auxin signalling is involved in nodule formation. Roy et al. extend this knowledge through the identification of a Medicago gene encoding MtLAX2 that is a functional homologue of the Arabidopsis auxin influx transporter AUX1. Loss of function mutants mtlax2 and aux1 show similar phenotypes including reduced sensitivity to auxin, reduced lateral root number, and reduced root gravitropism. Furthermore, MtLAX2 is expressed at very early stages of nodule formation and is required for nodule initiation. This study indicates that auxin influx is crucial for both nodule and lateral root organogenesis, and provides additional insights into the process of nodule formation. Plant Physiol. 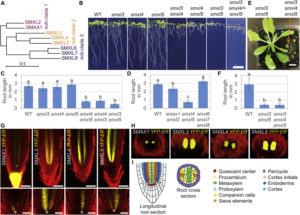 SMAX1 (SUPPRESSOR OF MAX2 1) was identified genetically as a suppressor of the enhanced seed dormancy phenotype of the max2 mutant, which is affected in both strigolactone (SL) and karrikin (KAR) responses (karrikins are smoke-derived germination promoters). SMAX1 is the founding member of an eight-member gene family in Arabidopsis, the SMAX1-LIKE family (SMXL); the encoded proteins show weak homology to HEAT SHOCK PROTEIN 101. Wallner et al. investigated the function of previously uncharacterized SMXL3, 4 and 5, through single mutants, double mutant combinations, and the triple mutant. The single mutants showed no abnormal phenotype, but all double mutants and the triple mutant showed reduced root growth. The genes are expressed in the protophloem, promote phloem development, and do not depend on SL or KAR signalling components. Curr. Biol.
SMAX1 (SUPPRESSOR OF MAX2 1) was identified genetically as a suppressor of the enhanced seed dormancy phenotype of the max2 mutant, which is affected in both strigolactone (SL) and karrikin (KAR) responses (karrikins are smoke-derived germination promoters). SMAX1 is the founding member of an eight-member gene family in Arabidopsis, the SMAX1-LIKE family (SMXL); the encoded proteins show weak homology to HEAT SHOCK PROTEIN 101. Wallner et al. investigated the function of previously uncharacterized SMXL3, 4 and 5, through single mutants, double mutant combinations, and the triple mutant. The single mutants showed no abnormal phenotype, but all double mutants and the triple mutant showed reduced root growth. The genes are expressed in the protophloem, promote phloem development, and do not depend on SL or KAR signalling components. Curr. Biol.  The maize shoot comprises modular domains of leaf, subtending shoot tissue, and an intercalary meristem that sits just above the next leaf down. By contrast to eudicot shoots that form a ring of vascular bundles, maize shoot vascular bundles remain as separate bundles, forming a “disordered” atactostele. Tsuda et al. used a reverse-genetic approach to investigate the function of BELL1-like Homeobox (BLH) transcription factors in maize, and focused on two that are highly expressed in the shoot meristem, BLH12 and BLH14. Both proteins interact in vivo and in vitro with the KN1 transcription factor. The double mutant blh12/14 was much shorter than its siblings and showed a premature differentiation of the intercalary meristem. Furthermore, in the double mutant vascular bundle number was significantly reduced due to both a decrease in their formation and an increase on their tendency to merge together (anastamose). This study then shows a role for BLH12 and BLH14 in preventing vein fusion, contributing to the maintenance of distinct vascular bundles in maize, and in preventing premature differentiation of the intercalary meristem. Plant Cell
The maize shoot comprises modular domains of leaf, subtending shoot tissue, and an intercalary meristem that sits just above the next leaf down. By contrast to eudicot shoots that form a ring of vascular bundles, maize shoot vascular bundles remain as separate bundles, forming a “disordered” atactostele. Tsuda et al. used a reverse-genetic approach to investigate the function of BELL1-like Homeobox (BLH) transcription factors in maize, and focused on two that are highly expressed in the shoot meristem, BLH12 and BLH14. Both proteins interact in vivo and in vitro with the KN1 transcription factor. The double mutant blh12/14 was much shorter than its siblings and showed a premature differentiation of the intercalary meristem. Furthermore, in the double mutant vascular bundle number was significantly reduced due to both a decrease in their formation and an increase on their tendency to merge together (anastamose). This study then shows a role for BLH12 and BLH14 in preventing vein fusion, contributing to the maintenance of distinct vascular bundles in maize, and in preventing premature differentiation of the intercalary meristem. Plant Cell  Several transcription-factor encoding genes involved in the transition from vegetative to reproductive growth have been identified. One of these, LEAFY (LFY), is expressed at the flanks of the inflorescence meristem at the site of newly forming floral meristems; loss-of-function lfy mutants produce leaves instead of flowers. APETALA1 (AP1) and CAULIFLOWER (CAL) are related genes expressed very early in floral meristem development; loss-of-function of both genes leads to an overproliferation of inflorescence meristem tissue, forming an apex that resembles a cauliflower. Goslin et al. looked at gene expression in a plant in which LFY was activated by fusion to the glucocorticoid receptor (LFY-GR) in an ap1/cal mutant background. Specific LFY targets in the mutant and wild-type background were compared, and compared to results from an AP1-GR study, to examine the regulatory interplay between these transcription factors. Although both genetic and molecular evidence suggest that LFY and AP1/CAL act in a partially redundant manner in floral meristem specification, some of their gene targets show antagonistic regulation. The authors suggest a model in which LFY and AP1/CAL form an incoherent feedback loop; “by providing positive as well as negative inputs, LFY might ensure that flower development can commence only when AP1/CAL levels are sufficiently high to efficiently activate the expression of other key floral regulators.” Plant Physiol.
Several transcription-factor encoding genes involved in the transition from vegetative to reproductive growth have been identified. One of these, LEAFY (LFY), is expressed at the flanks of the inflorescence meristem at the site of newly forming floral meristems; loss-of-function lfy mutants produce leaves instead of flowers. APETALA1 (AP1) and CAULIFLOWER (CAL) are related genes expressed very early in floral meristem development; loss-of-function of both genes leads to an overproliferation of inflorescence meristem tissue, forming an apex that resembles a cauliflower. Goslin et al. looked at gene expression in a plant in which LFY was activated by fusion to the glucocorticoid receptor (LFY-GR) in an ap1/cal mutant background. Specific LFY targets in the mutant and wild-type background were compared, and compared to results from an AP1-GR study, to examine the regulatory interplay between these transcription factors. Although both genetic and molecular evidence suggest that LFY and AP1/CAL act in a partially redundant manner in floral meristem specification, some of their gene targets show antagonistic regulation. The authors suggest a model in which LFY and AP1/CAL form an incoherent feedback loop; “by providing positive as well as negative inputs, LFY might ensure that flower development can commence only when AP1/CAL levels are sufficiently high to efficiently activate the expression of other key floral regulators.” Plant Physiol. 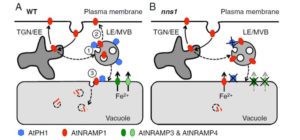 NRAMPs are transporters of iron (Fe) and manganese (Mn). The nramp3nramp4 double mutant arrests shortly after germination, due to its inability to remobilize Fe from seed vacuolar stores. Agorio et al. used a genetic approach to identify nns1, a partial suppressor of the growth-arrest phenotype. They mapped nns1 to AtPH1, a gene encoding a pleckstrin-homology (PH) domain protein. The PH domain binds the membrane lipid phosphatidylinositol 3-phosphate (PI3P) – PI3P and other phosphorylated PIs are known to specifically recruit proteins to membrane subdomains. In the nns1 mutant that lacks AtPH1, NRAMP1 (normally found in the endosomal pathway and at the plasma membrane) is mislocalized to the vacuole, where it is thought to complement the missing iron transport function of the nramp3nramp4 mutant. Proc. Natl. Acad. Sci. USA
NRAMPs are transporters of iron (Fe) and manganese (Mn). The nramp3nramp4 double mutant arrests shortly after germination, due to its inability to remobilize Fe from seed vacuolar stores. Agorio et al. used a genetic approach to identify nns1, a partial suppressor of the growth-arrest phenotype. They mapped nns1 to AtPH1, a gene encoding a pleckstrin-homology (PH) domain protein. The PH domain binds the membrane lipid phosphatidylinositol 3-phosphate (PI3P) – PI3P and other phosphorylated PIs are known to specifically recruit proteins to membrane subdomains. In the nns1 mutant that lacks AtPH1, NRAMP1 (normally found in the endosomal pathway and at the plasma membrane) is mislocalized to the vacuole, where it is thought to complement the missing iron transport function of the nramp3nramp4 mutant. Proc. Natl. Acad. Sci. USA