Striga hermonthica – a beautiful but devastating plant…
This week’s post was written by Caroline Wood, a PhD candidate at the University of Sheffield.
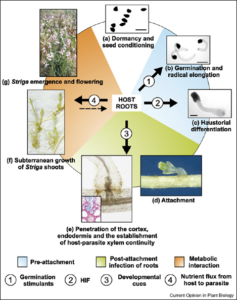 When it comes to crop diseases, insects, viruses, and fungi may get the media limelight but in certain regions it is actually other plants which are a farmer’s greatest enemy. In sub-Saharan Africa, one weed in particular – Striga hermonthica – is an almost unstoppable scourge and one of the main limiting factors for food security.
When it comes to crop diseases, insects, viruses, and fungi may get the media limelight but in certain regions it is actually other plants which are a farmer’s greatest enemy. In sub-Saharan Africa, one weed in particular – Striga hermonthica – is an almost unstoppable scourge and one of the main limiting factors for food security.
Striga is a parasitic plant; it attaches to and feeds off a host plant. For most of us, parasitic plants are simply harmless curiosities. Over 4,000 plants are known to have adopted a parasitic mode of life, including the seasonal favorite mistletoe (a stem parasite of conifers) and Rafflesia arnoldii, nicknamed the “corpse flower” for its huge, smelly blooms. Although the latter produces the world’s largest flower, it has no true roots – only thread-like structures that infect tropical vines.
Click here to read the rest of this post on the Global Plant Council blog.


 “As I am coming to the end of my time as postdoc I have been thinking a lot about what I have learnt and perhaps wish I knew when I started out. Inspired by the German poet
“As I am coming to the end of my time as postdoc I have been thinking a lot about what I have learnt and perhaps wish I knew when I started out. Inspired by the German poet 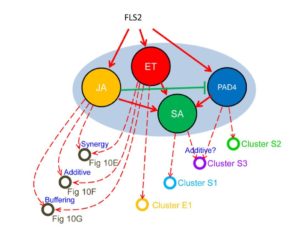 Classic genetic approaches have been instrumental in identifying genes that control developmental and defense networks, but as Hillmer et al. point out, analysis of single mutants is complicated by network buffering. They describe network buffering as occurring when the effect of losing a gene’s function is masked by the function of other genes. They point out that the immune signaling network must be highly buffered as it is constantly under attack by pathogens. Previously, they defined the immune network as being made up of four sub-networks, each of which can be interrupted by disabling a single gene. Specifically, they can eliminate the jasmonate, salicylate, ethylene and PAD4 sectors by elimination of a biosynthetic or signaling gene. They previously published an analysis of higher order mutants knocking out all combinations of these sub-networks. Here they analyze transcriptomic and hormone profiles of each of these mutants in response to flg22, a peptide that mimics the presence of a pathogen. Their results demonstrate that the immune network “is not a collection of independent pathways; rather interactions among the sectors dominate network regulation of the transcriptome response.” PLOS Genet.
Classic genetic approaches have been instrumental in identifying genes that control developmental and defense networks, but as Hillmer et al. point out, analysis of single mutants is complicated by network buffering. They describe network buffering as occurring when the effect of losing a gene’s function is masked by the function of other genes. They point out that the immune signaling network must be highly buffered as it is constantly under attack by pathogens. Previously, they defined the immune network as being made up of four sub-networks, each of which can be interrupted by disabling a single gene. Specifically, they can eliminate the jasmonate, salicylate, ethylene and PAD4 sectors by elimination of a biosynthetic or signaling gene. They previously published an analysis of higher order mutants knocking out all combinations of these sub-networks. Here they analyze transcriptomic and hormone profiles of each of these mutants in response to flg22, a peptide that mimics the presence of a pathogen. Their results demonstrate that the immune network “is not a collection of independent pathways; rather interactions among the sectors dominate network regulation of the transcriptome response.” PLOS Genet. The interaction between wild tobacco (Nicotiana attenuata) and the moth Manduca sexta, which is both a pollinator and a herbivore, is a model for plant/ arthropod interactions and has revealed insights into chemical signals and defenses. Zhou et al. show that a single compound, the sesquiterpene (E)-α-bergamotene, serves as both a pollinator attractor when produced in flowers at night (when the adult moths pollinators are active), and when produced in leaves acts as an indirect defense signal by attracting predators, which eat the herbivorous larvae. Interestingly, a single gene that is under circadian control is responsible for the production of this compound in flowers and leaves. Curr. Biol.
The interaction between wild tobacco (Nicotiana attenuata) and the moth Manduca sexta, which is both a pollinator and a herbivore, is a model for plant/ arthropod interactions and has revealed insights into chemical signals and defenses. Zhou et al. show that a single compound, the sesquiterpene (E)-α-bergamotene, serves as both a pollinator attractor when produced in flowers at night (when the adult moths pollinators are active), and when produced in leaves acts as an indirect defense signal by attracting predators, which eat the herbivorous larvae. Interestingly, a single gene that is under circadian control is responsible for the production of this compound in flowers and leaves. Curr. Biol. 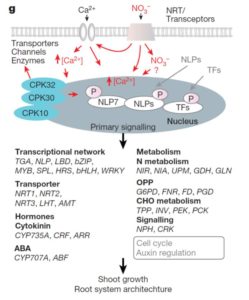 Nitrate acts as a potent signal as well as a source of nutritional nitrogen, but key players in the nitrate response have been missing from our understanding. Liu et al. identified a unique calcium signal stimulated by nitrate in mesophyll cells. They then found that in vitro kinase activity of Ca2+-sensor protein kinases (CPKs) are also nitrate stimulated, and they assayed constitutively-active Arabidopsis CPKs for stimulation of a nitrate-responsive reporter in response to nitrate. They found that several CPK genes act synergistically in the nitrate response. Efforts to generate higher-order CPK mutants were thwarted by embryo lethality, so the authors generated inducible mutants. The ipck mutant is a triple-inducible mutant of CPK10, CPK30 and CPK32. It shows significant defects in nitrate-responsive transcription and growth, and phosphorylation of NLP transcription factors. These results reveal a new Ca2+-CPK-NLP signaling cascade mediating nitrate responses. Nature
Nitrate acts as a potent signal as well as a source of nutritional nitrogen, but key players in the nitrate response have been missing from our understanding. Liu et al. identified a unique calcium signal stimulated by nitrate in mesophyll cells. They then found that in vitro kinase activity of Ca2+-sensor protein kinases (CPKs) are also nitrate stimulated, and they assayed constitutively-active Arabidopsis CPKs for stimulation of a nitrate-responsive reporter in response to nitrate. They found that several CPK genes act synergistically in the nitrate response. Efforts to generate higher-order CPK mutants were thwarted by embryo lethality, so the authors generated inducible mutants. The ipck mutant is a triple-inducible mutant of CPK10, CPK30 and CPK32. It shows significant defects in nitrate-responsive transcription and growth, and phosphorylation of NLP transcription factors. These results reveal a new Ca2+-CPK-NLP signaling cascade mediating nitrate responses. Nature 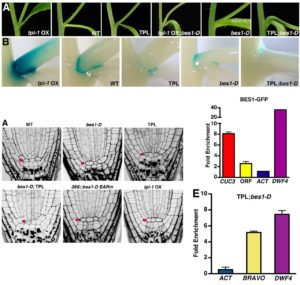 Shoot and root development in Arabidopsis thaliana are controlled by many factors, one of which is brassinosteroid (BR) signaling. BR-responsive gene expression is controlled by the BES1 and BZR1 transcription factors. Using its EAR domain, BES1 binds to the transcriptional repressor TOPLESS (TPL) to activate and repress genes. Espinosa-Ruiz et al. show genetic evidence for interaction between BES1 and TPL by using the bes1-D mutant that shows a constitutive BR response. Expression of the tpl-1 mutant reverses the bes1-D phenotype. CUP-SHAPED COTYLEDON (CUC) genes mark organ boundaries, and both TPL and BES1 repress CUC3 to separate organs. In the root quiescent center, cell division is tightly regulated. TPL binds with BES1 in the quiescent center to regulate the division repressor BRAVO. The BES1-TPL transcription regulation unit controls signals for cell proliferation in the root and shoot. (Summary by
Shoot and root development in Arabidopsis thaliana are controlled by many factors, one of which is brassinosteroid (BR) signaling. BR-responsive gene expression is controlled by the BES1 and BZR1 transcription factors. Using its EAR domain, BES1 binds to the transcriptional repressor TOPLESS (TPL) to activate and repress genes. Espinosa-Ruiz et al. show genetic evidence for interaction between BES1 and TPL by using the bes1-D mutant that shows a constitutive BR response. Expression of the tpl-1 mutant reverses the bes1-D phenotype. CUP-SHAPED COTYLEDON (CUC) genes mark organ boundaries, and both TPL and BES1 repress CUC3 to separate organs. In the root quiescent center, cell division is tightly regulated. TPL binds with BES1 in the quiescent center to regulate the division repressor BRAVO. The BES1-TPL transcription regulation unit controls signals for cell proliferation in the root and shoot. (Summary by 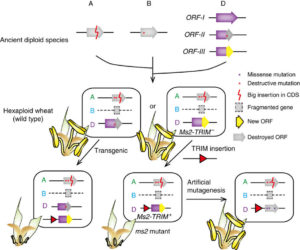 The first dominant male-sterile mutant in wheat, ms2 was identified more than 40 years ago, and Xia et al. have now identified the molecular basis for this trait. They find that the mutation is caused by a transposon (a terminal-repeat retrotransposon in miniature, or TRIM) insertion into the promoter of the Ms2 gene, causing it to be specifically expressed in anthers. The protein encoded by the Ms2 gene has no known functional domains, and only has homologues in the diploid ancestor of wheat, and tetraploid and hexaploid wheat, but functional analysis indicates that it is the cause of the male-sterile phenotype. The authors summarize, “The identification of Ms2 not only unravels the genetic basis of a historically important breeding trait, but also shows an example of how a TRIM element insertion near a gene can contribute to genetic novelty and phenotypic plasticity.” Nature Comms.
The first dominant male-sterile mutant in wheat, ms2 was identified more than 40 years ago, and Xia et al. have now identified the molecular basis for this trait. They find that the mutation is caused by a transposon (a terminal-repeat retrotransposon in miniature, or TRIM) insertion into the promoter of the Ms2 gene, causing it to be specifically expressed in anthers. The protein encoded by the Ms2 gene has no known functional domains, and only has homologues in the diploid ancestor of wheat, and tetraploid and hexaploid wheat, but functional analysis indicates that it is the cause of the male-sterile phenotype. The authors summarize, “The identification of Ms2 not only unravels the genetic basis of a historically important breeding trait, but also shows an example of how a TRIM element insertion near a gene can contribute to genetic novelty and phenotypic plasticity.” Nature Comms. 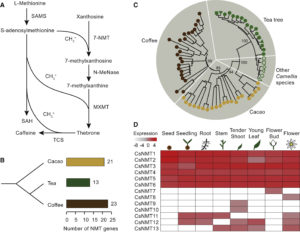 The release of the tea (Camellia sinensis) genome completes the set of the three major caffeine-producing plants (also coffee and cacao). (Note that the title of this article refers to the “tea tree”, a name that is often used for a different plant, Melaleuca alternifolia, the source of tea tree oil; the genome described here is of the tea beverage plant). Key findings include an expansion of genes involved in the synthesis of flavonoids that contribute to tea flavor. The data also show that caffeine synthesis evolved separately in tea as from coffee and cacao. Xia et al. point out that this is the first genome in the genus Camellia, which of course includes horticultural as well as oil-bearing species. Mol. Plant
The release of the tea (Camellia sinensis) genome completes the set of the three major caffeine-producing plants (also coffee and cacao). (Note that the title of this article refers to the “tea tree”, a name that is often used for a different plant, Melaleuca alternifolia, the source of tea tree oil; the genome described here is of the tea beverage plant). Key findings include an expansion of genes involved in the synthesis of flavonoids that contribute to tea flavor. The data also show that caffeine synthesis evolved separately in tea as from coffee and cacao. Xia et al. point out that this is the first genome in the genus Camellia, which of course includes horticultural as well as oil-bearing species. Mol. Plant  An old ad stated, “Without chemicals, life itself would be impossible,” but it’s only more recently that we’ve begun to understand the importance of semiochemicals – chemicals produced for communication. Leach et al. pull together insights from chemical ecologists, soil scientists, plant pathologists and entomologists in this excellent review of the phytobiome (all the living organisms in and around a plant) and the communication within it. They start by estimating the number and diversity of organisms (viruses, bacteria, fungi, arthropods, nematodes, protists and earthworms) that live together in the phytobiome, and then what we understand about their chemical signals. The authors conclude, “The nascent field of phytobiomes research is an attempt to strengthen the connections between ecological and agricultural bodies of knowledge for better translation to crop and managed ecosystems”, and direct readers to the
An old ad stated, “Without chemicals, life itself would be impossible,” but it’s only more recently that we’ve begun to understand the importance of semiochemicals – chemicals produced for communication. Leach et al. pull together insights from chemical ecologists, soil scientists, plant pathologists and entomologists in this excellent review of the phytobiome (all the living organisms in and around a plant) and the communication within it. They start by estimating the number and diversity of organisms (viruses, bacteria, fungi, arthropods, nematodes, protists and earthworms) that live together in the phytobiome, and then what we understand about their chemical signals. The authors conclude, “The nascent field of phytobiomes research is an attempt to strengthen the connections between ecological and agricultural bodies of knowledge for better translation to crop and managed ecosystems”, and direct readers to the