All in good time: Timing gene activation during early flower development
Pelayo et al. explore the function of a biological timer that activates key genes for flowering.
Margaret Anne Pelayo, Nara Institute of Science and Technology
https://doi.org/10.1093/plcell/koad123
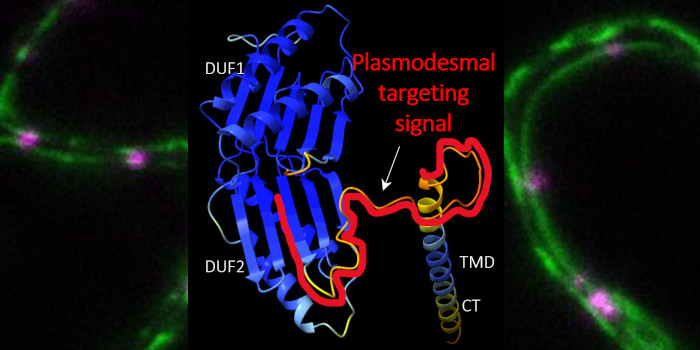
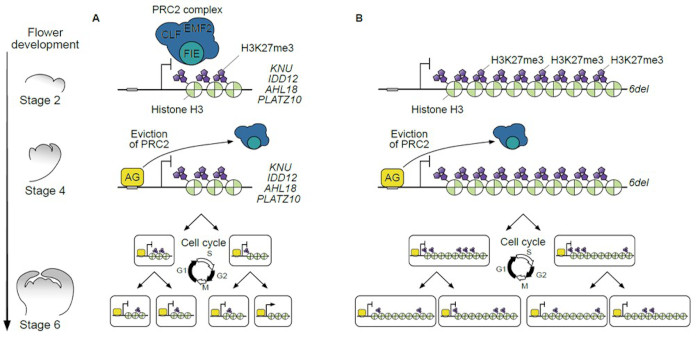
Background: For flowers to form, the floral meristem (floral stem cells) must irreversibly commit to becoming cells making up the various floral organs (sepals, petals, stamens, and carpels), a process known as floral meristem termination. Proper timing of floral meristem termination involves temporal activation of the transcription factor gene KNUCKLES (KNU) by its upstream regulator AGAMOUS (AG) via cell cycle-dependent dilution of the repressive histone modification at lysine 27 of histone H3 (H3K27me3) along the KNU coding sequence. This intrinsic ‘biotimer’ will activate KNU at precisely the right time to ensure proper flower development.
Question: Are there other genes similarly regulated by AGAMOUS during flower development making up a biotimer transcriptional regulatory network and can we manipulate KNU gene activation timing based on the set of criteria that defines the biotimer mechanism?
Findings: Using the model plant Arabidopsis thaliana, we found that the genes AT HOOK MOTIF NUCLEAR LOCALIZED PROTEIN18 (AHL18) and PLATZ10 are direct biotimer-regulated AG targets among a set of 23 biotimer candidate genes (including KNU). AHL18 and PLATZ10 are likely involved during stamen development, specifically for proper stamen elongation and maturation, respectively. We also introduced a simple mathematical model correlating the length of H3K27me3-marked regions at biotimer genes with gene activation timing and correctly predicted the timing of activation for KNU, AHL18, and PZ10. We validated the model’s predictions experimentally by modifying KNU gene length with tandem repeats of a H3K27me3-dense region in KNU’s coding sequence named del resulting in delayed and reduced KNU expression in a PRC2- and cell cycle–dependent manner.
Next steps: Our current work provides insight into epigenetic approaches for tunable gene expression and provides a mechanistic framework to understand which aspects of transcriptional regulatory systems can be effectively manipulated for future work aimed at enhancing plant productivity and resilience.
Reference:
Margaret Anne Pelayo, Fumi Morishita, Haruka Sawada, Kasumi Matsushita, Hideaki Iimura, Zemiao He, Liang Sheng Looi, Naoya Katagiri, Asumi Nagamori, Takamasa Suzuki, Marek Širl, Aleš Soukup, Akiko Satake, Toshiro Ito and Nobutoshi Yamaguchi (2023) AGAMOUS regulates various target genes via cell cycle–coupled H3K27me3 dilution in floral meristems and stamens. https://doi.org/10.1093/plcell/koad123


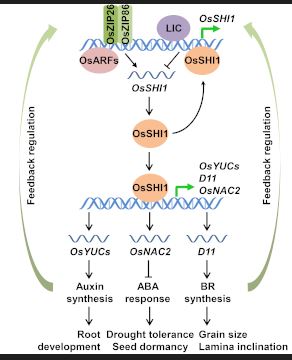 Findings: We found that the shi1 mutant exhibits many hormone-related morphological variations, such as auxin and brassinosteroid (BR)-regulated plant development as well as abscisic acid (ABA)-mediated stress tolerance. Further functional analysis revealed that several upstream transcription factors of auxin, ABA and BR pathways bind to the promoter region of OsSHI1 to activate or inhibit its transcription. OsSHI1 protein then directly regulates several auxin and BR biosynthesis genes, thus integrating the biosynthesis of auxin and BR with ABA signal transduction to coordinate plant growth and stress adaptation. In addition, OsSHI1 also negatively regulates its own expression. These findings demonstrate that OsSHI1 is a key transcriptional hub for the integration of multiple hormone signaling pathways.
Findings: We found that the shi1 mutant exhibits many hormone-related morphological variations, such as auxin and brassinosteroid (BR)-regulated plant development as well as abscisic acid (ABA)-mediated stress tolerance. Further functional analysis revealed that several upstream transcription factors of auxin, ABA and BR pathways bind to the promoter region of OsSHI1 to activate or inhibit its transcription. OsSHI1 protein then directly regulates several auxin and BR biosynthesis genes, thus integrating the biosynthesis of auxin and BR with ABA signal transduction to coordinate plant growth and stress adaptation. In addition, OsSHI1 also negatively regulates its own expression. These findings demonstrate that OsSHI1 is a key transcriptional hub for the integration of multiple hormone signaling pathways.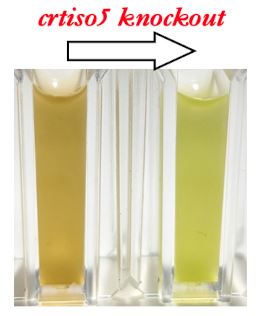 Findings: We found that a protein predicted to be from a family of carotenoid isomerases (CRTISO5) has gained a completely novel function in the fucoxanthin-containing diatom Phaeodactylum. When this gene is disrupted using clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated genome editing, the normally brown diatom appears green and shows a reduced light-harvesting capability and growth. We used a combination of genetics, enzyme assays and molecular structural analyses to show that CRTISO5 catalyzes the final step of fucoxanthin biosynthesis. Surprisingly, instead of serving as an isomerase, it hydrates a carbon-carbon triple bond within the precursor molecule, leading to formation of the characteristic keto group in fucoxanthin.
Findings: We found that a protein predicted to be from a family of carotenoid isomerases (CRTISO5) has gained a completely novel function in the fucoxanthin-containing diatom Phaeodactylum. When this gene is disrupted using clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated genome editing, the normally brown diatom appears green and shows a reduced light-harvesting capability and growth. We used a combination of genetics, enzyme assays and molecular structural analyses to show that CRTISO5 catalyzes the final step of fucoxanthin biosynthesis. Surprisingly, instead of serving as an isomerase, it hydrates a carbon-carbon triple bond within the precursor molecule, leading to formation of the characteristic keto group in fucoxanthin.