The phosphatase PC1 switches off catalase to balance salt tolerance and growth
Cong Liu, Jianzhong Lin, and Xuanming Liu and colleagues show that the protein phosphatase PC1 dephosphorylates and deactivates CatC to negatively regulate H2O2 homeostasis and salt tolerance in rice.
https://doi.org/10.1093/plcell/koad167
By Cong Liu, Jianzhong Lin, and Xuanming Liu
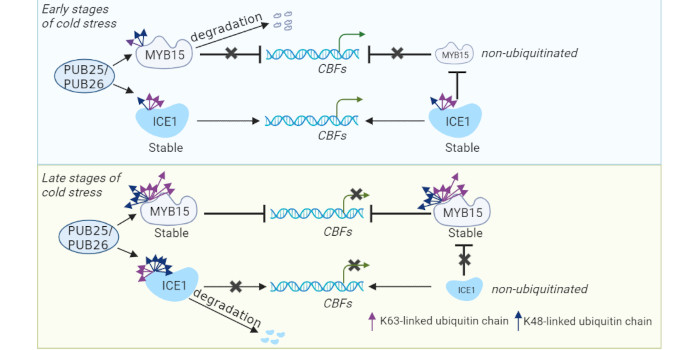
Background: Soil salinity is a worldwide problem that threatens the growth and yield of crops and prevents the sustainable development of modern agriculture. Catalase is a type of hydrogen peroxide (H2O2)–scavenging enzyme that plays a central role in stress responses as well as growth and development. Catalase is a phosphoprotein whose function can be tightly regulated by phosphorylation. Several catalase kinases have been extensively studied, but its phosphatase remains unclear.
Question: How is catalase switched off by phosphatases to balance stress response and growth?
Findings: We identified a protein phosphatase of the catalase CatC (PC1) that dephosphorylates CatC at the Ser-9 residue in rice. Upon salt stress, PC1 is inhibited and leaves the catalase tetramer intact, an oligomeric form essential for high catalase activity, leading to improved salt tolerance. Once salt stress is alleviated, PC1 is activated and dephosphorylates CatC at Ser-9 to accelerate its disassociation to monomers, thereby keeping an appropriate H2O2 level to sustain rice growth and development. Thus, PC1 plays an important role during the transition from salt stress to normal growth conditions. Moreover, genetic manipulation of PC1 can limit yield loss in rice grain under salt stress.
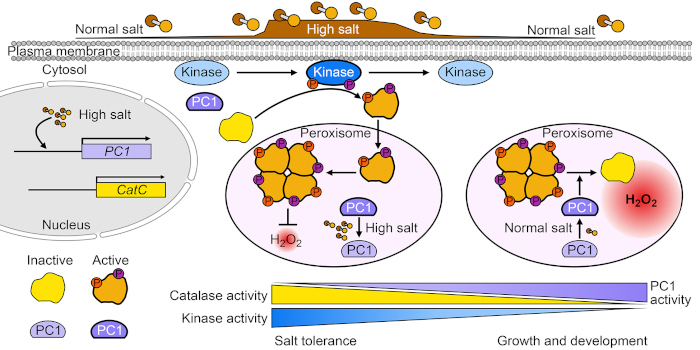
Next steps: The functional analysis of PC1 sheds light on the switch-off mechanisms of CAT in plants. The kinase that phosphorylates CatC at Ser-9 and how PC1 is activated and inhibited require further investigation.
Reference:
Cong Liu, Jian-Zhong Lin, Yan Wang, Ye Tian, He-Ping Zheng, Zheng-Kun Zhou, Yan-Biao Zhou, Xiao-Dan Tang, Xin-Hui Zhao, Ting Wu, Shi-Long Xu, Dong-Ying Tang, Ze-Cheng Zuo, Hang He, Lian-Yang Bai, Yuan-Zhu Yang and Xuan-Ming Liu The Protein Phosphatase PC1 Dephosphorylates and Deactivates CatC to Negatively Regulate H2O2 Homeostasis and Salt Tolerance in Rice (2023) https://doi.org/10.1093/plcell/koad167


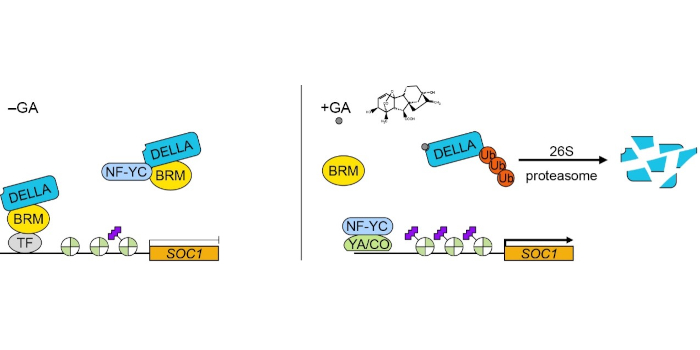 Next steps: We will investigate whether the BRM-NF-Y module is also responsive to other phytohormone signals and how BRM integrates different phytohormone signals during plant development.
Next steps: We will investigate whether the BRM-NF-Y module is also responsive to other phytohormone signals and how BRM integrates different phytohormone signals during plant development.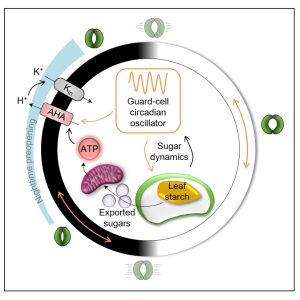 Findings: We developed PhenoLeaks, a phenotyping pipeline to analyze the transpiration dynamics over days and nights on 150 plants at the same time. We screened a collection of Arabidopsis starch mutants and found that severe mutations in starch metabolism not only disrupt stomatal preopening at night, but also delay endogenous stomatal movements during the whole day. When the lesions in starch metabolism were confined to the guard cells, stomata showed normal endogenous movements, suggesting that starch from the rest of the leaf is able to set the tempo of stomatal movements, most likely by providing sugars that interact with the guard-cell circadian clock.
Findings: We developed PhenoLeaks, a phenotyping pipeline to analyze the transpiration dynamics over days and nights on 150 plants at the same time. We screened a collection of Arabidopsis starch mutants and found that severe mutations in starch metabolism not only disrupt stomatal preopening at night, but also delay endogenous stomatal movements during the whole day. When the lesions in starch metabolism were confined to the guard cells, stomata showed normal endogenous movements, suggesting that starch from the rest of the leaf is able to set the tempo of stomatal movements, most likely by providing sugars that interact with the guard-cell circadian clock.