Xu et al. demonstrate the transcriptional regulatory steps responsible for establishing the regenerating capability of callus during Arabidopsis regeneration.
https://doi.org/10.1093/plcell/koad255
By Chongyi Xu1, Pengjie Chang1,2, Shiqi Guo1,2, Xiaona Yang1,2, Xinchun Liu1,2, Baofeng Sui1,2, Dongxue Yu1,2, Wei Xin1,2, and Yuxin Hu1,3
1 Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, China National Botanical Garden, Beijing 100093, China.
2 University of Chinese Academy of Sciences, Beijing 100049, China.
3 National Center for Plant Gene Research, Beijing 100093, China.
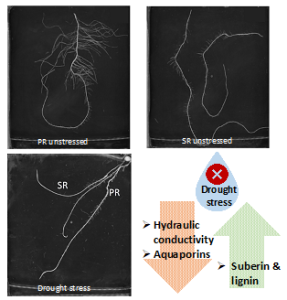
Background: Plant cells retain a remarkable capacity to regenerate new organs or entire individuals in the real world and under tissue-culture conditions. A well-established in vitro plant regeneration procedure applicable for transgenic and biotechnological uses generally starts with the induction of pluripotent callus cells, which is required for subsequent de novo shoot or root regeneration. Recent studies in Arabidopsis (Arabidopsis thaliana) have revealed that auxin-induced ectopic activation of root stem cell factors within callus cells establishes the shoot-regenerating capability, and some auxin signaling components involved in root development, such as AUXIN RESPONSE FACTOR (ARFs) and their downstream transcription factors LATERAL ORGAN BOUNDARIES DOMAIN (LBD), play critical roles in directing callus formation. However, the molecular link between these auxin signaling components and activation of root stem cell factors during callus induction is missing.
Question: What are the factors responsible for activation of root stem cell factors to establish callus pluripotency in Arabidopsis for in vitro regeneration?
Findings: We identified the Arabidopsis transcription factors WRKY23 and bHLH041 as a transcriptional activator and repressor, respectively, of the expression of root stem cell genes during auxin-induced callus formation. Genetic and molecular evidence revealed that auxin-induced WRKY23 downstream of ARF7 and ARF19 directly activates the transcription of PLETHORA 3 (PLT3) and PLT7 and the downstream target genes of their encoded proteins PLT1, PLT2, and WUSCHEL-RELATED HOMEOBOX 5 (WOX5), while LBD induces the removal of bHLH041, alleviating the transcriptional repression of PLT1, PLT2, and WOX5. We also demonstrated that two transcriptional pathways synergize the shoot-regenerating capability of callus cells. These findings elucidate the transcriptional mechanism underlying callus pluripotency establishment, which links auxin signaling and cellular reprogramming during in vitro plant regeneration programs.
Next steps: It will be worth identifying the orthologs of WRKY23 and bHLH041 in crops and economically important plants and exploring whether such regulatory mechanisms are conserved, which would potentially boost regeneration-based transgene and gene editing in these species.
Reference:
Chongyi Xu, Pengjie Chang, Shiqi Guo, Xiaona Yang, Xinchun Liu, Baofeng Sui, Dongxue Yu, Wei Xin, Yuxin Hu (2023). Transcriptional activation by WRKY23 and derepression by removal of bHLH041 coordinately establish callus pluripotency in Arabidopsis regeneration. https://doi.org/10.1093/plcell/koad255
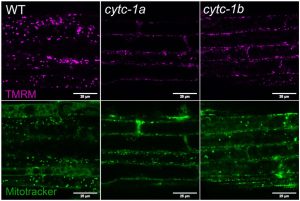 As seedlings grow, they continuously fine-tune the balance between plant growth and energy consumption for better plant fitness. Mitochondria are essential to generate energy. Here, Canal et al. generated single mutants for a gene involved in the mitochondrial respiratory chain – CYTOCHROME C-1 (CYTC-1), which caused defective mitochondrial energy production, membrane potential, lower ATP content, and a reduced growth rate in the cytc-1 mutants. Some of the mutant phenotypes resemble those of plants with abnormal Target of Rapamycin (TOR) signaling. TOR is a protein that integrates growth-related processes in all eukaryotes. The authors showed that TOR activity is impaired in the cytc-1 mutants. In contrast, TOR overexpression rescued the seedling growth and low ATP level of cytc-1 mutant. The authors conclude that plants can coordinate their metabolism in response to the disruption of the mitochondrial function in the cytc-1 mutation, which anticipates energy exhaustion and speculates that conditions by reducing TOR activity levels and slowing growth. This study also sheds light on how mitochondria communicate their status to the rest of the cell. (Summary by Yueh Cho @YuehCho1984) New Phytol. 10.1111/nph.19506.
As seedlings grow, they continuously fine-tune the balance between plant growth and energy consumption for better plant fitness. Mitochondria are essential to generate energy. Here, Canal et al. generated single mutants for a gene involved in the mitochondrial respiratory chain – CYTOCHROME C-1 (CYTC-1), which caused defective mitochondrial energy production, membrane potential, lower ATP content, and a reduced growth rate in the cytc-1 mutants. Some of the mutant phenotypes resemble those of plants with abnormal Target of Rapamycin (TOR) signaling. TOR is a protein that integrates growth-related processes in all eukaryotes. The authors showed that TOR activity is impaired in the cytc-1 mutants. In contrast, TOR overexpression rescued the seedling growth and low ATP level of cytc-1 mutant. The authors conclude that plants can coordinate their metabolism in response to the disruption of the mitochondrial function in the cytc-1 mutation, which anticipates energy exhaustion and speculates that conditions by reducing TOR activity levels and slowing growth. This study also sheds light on how mitochondria communicate their status to the rest of the cell. (Summary by Yueh Cho @YuehCho1984) New Phytol. 10.1111/nph.19506.