On UPF Proteins, Baking Cookies, and the Many Targets of Nonsense-Mediated RNA Decay
When we cut out cookies from rolled-out dough, we often end up with unwanted dough scraps, and sometimes the dough sticks to the cookie-cutter, resulting in misshapen cookies. Just like these misshapen cookies, faulty or aberrant RNAs can arise in cells due to mistakes during transcription, RNA processing, or frameshift mutations. Nonsense-mediated decay (NMD) is a major eukaryotic RNA surveillance pathway that rapidly degrades aberrant RNAs and thus prevents the accumulation of truncated, dysfunctional proteins––like having a little helper in the kitchen to gobble up the scraps of cookie dough. NMD also controls the turnover of many normal or naturally occurring RNAs. Notably, NMD fine-tunes the expression of many R genes that function in plant immunity (Jung et al. 2020). Impaired NMD leads to enhanced immune response as well as the deregulation of several developmental processes.
The core NMD machinery consists of three evolutionarily conserved UP FRAMESHIFT (UPF) proteins: UPF1, UPF2, and UPF3. The assembly of the NMD machinery is promoted by the recognition of transcripts with premature translation termination codons or long 3′ UTRs. This leads to UPF1 activation, which blocks translation initiation. Activated UPF1 also recruits SUPPRESSOR WITH MORPHOGENETIC EFFECT ON GENITALIA 7 (SMG7), which results in RNA degradation via deadenylation and decapping. Although UPF1, UPF3, and SMG7 all encode NMD components, loss-of-function mutants in these genes exhibit distinct phenotypes in Arabidopsis thaliana. For example, upf1 mutants are seedling lethal due to the strong activation of the immune response, but upf3 mutants are viable, with mild growth defects and abnormal flowers. The smg7 mutants are also viable but have a more severe growth retardation phenotype than that of upf3 (Capitao et al. 2018).
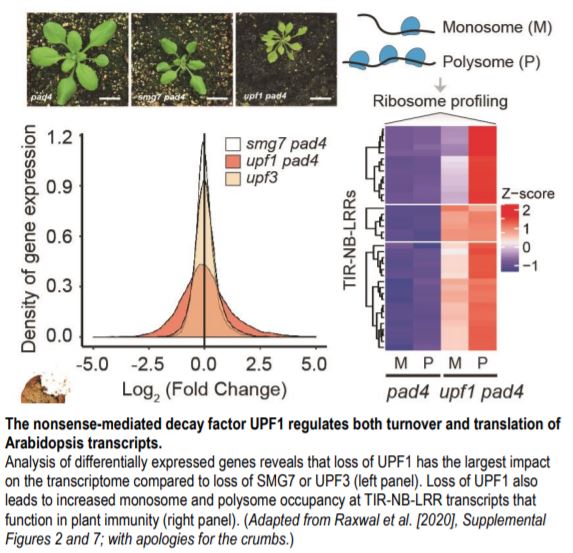 Why do NMD mutants show such distinct phenotypes? A recent study by Raxwal et al. (2020) tackled this question by comparing transcriptome-wide gene expression patterns of viable upf1-3 pad4 and smg7-1 pad4 double mutants in Arabidopsis. These loss-of-function NMD mutants have an attenuated immune response due to the inactivation of PHYTOALEXIN DEFICIENT 4 (PAD4). Therefore, these mutants provide a better resolution of NMD targets from transcripts that are upregulated due to defense activation. The authors found that upf1-3 pad4 mutants show the highest accumulation of known aberrant transcripts harboring NMD-inducing features compared to the other NMD factors. The loss of UPF1 significantly affects ~21% of the transcriptome, but loss of SMG7 affects only ~4% (see figure). The loss of UPF3 affects an even smaller fraction of the transcriptome (<100 genes), consistent with its mild mutant phenotype.
Why do NMD mutants show such distinct phenotypes? A recent study by Raxwal et al. (2020) tackled this question by comparing transcriptome-wide gene expression patterns of viable upf1-3 pad4 and smg7-1 pad4 double mutants in Arabidopsis. These loss-of-function NMD mutants have an attenuated immune response due to the inactivation of PHYTOALEXIN DEFICIENT 4 (PAD4). Therefore, these mutants provide a better resolution of NMD targets from transcripts that are upregulated due to defense activation. The authors found that upf1-3 pad4 mutants show the highest accumulation of known aberrant transcripts harboring NMD-inducing features compared to the other NMD factors. The loss of UPF1 significantly affects ~21% of the transcriptome, but loss of SMG7 affects only ~4% (see figure). The loss of UPF3 affects an even smaller fraction of the transcriptome (<100 genes), consistent with its mild mutant phenotype.
Aberrant RNA accumulation in NMD mutants usually manifests as upregulation of the associated genes and both smg7-1 pad4 and upf3-1 mutants show upregulation of more than 80% of the differentially expressed genes. Surprisingly, only ~55% of the differentially expressed genes in upf1 pad4 are upregulated, suggesting that UPF1 has a broad impact on the transcriptome beyond its role in aberrant RNA decay. Gene ontology analysis further illustrates the distinct characteristics of genes regulated by different NMD factors. For instance, only upf1-3 pad4 mutants show significant upregulation of genes involved in alternative splicing. Reassembly of the upf1-3 pad4 transcriptome also revealed widespread changes in alternative splicing. These findings confirm that UPF1 plays an important role in splicing regulation.
Interestingly, upf1-3 pad4 mutants significantly downregulate processes related to translation and ribosome biogenesis. Actively translated RNAs are usually bound to multiple ribosomes (i.e. polysomes), whereas poorly translated RNAs are bound to single ribosomes (monosomes). The authors found that in the upf1-3 pad4 mutants, transcripts showed a small but significant shift from polysomes to monosomes, suggesting that UPF1 is important for normal mRNA translation. At the same time, rare and less stable transcripts in upf1-3 pad4 show increased polysome occupancy compared to the bulk transcriptome. Loss of UPF1 also leads to increased translation of TIR-NB-LRR-type R genes that are well-known targets of NMD (see figure). Hence, UPF1 affects the transcript levels as well as the translation of NMD targets.
The findings of Raxwal et al. (2020) emphasize a bigger role for UPF1 in regulating naturally occurring transcripts than previously thought––as if your little helper was cleaning up cookie dough and taking out the trash. Future research (along with more cookies) is required to understand how UPF1 recognizes such a diverse set of targets and triggers their degradation, even in the absence of a functional SMG7.
Saima Shahid
Donald Danforth Plant Science Center
Saint Louis, Missouri
ORCID: 0000-0001-9385-0925
REFERENCES
Jung, H.W., Panigrahi, G.K., Jung, G.Y., Lee, Y.J., Shin, K.H., Sahoo, A., Choi, E.S., Lee, E., Kim, K.M., Yang, S.H. and Jeon, J.S. (2020). Pathogen-associated molecular pattern-triggered immunity involves proteolytic degradation of core nonsense-mediated mRNA decay factors during the early defense response. Plant Cell 32: 1081-1101.
Capitao, C., Shukla, N., Wandrolova, A., Mittelsten Scheid, O., and Riha, K. (2018). Functional characterization of SMG7 paralogs in Arabidopsis thaliana. Front. Plant Sci. 9: 1602.