Non-immunogenic bacterial epitopes mask recognition of their immunogenic counterparts
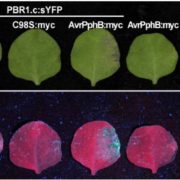
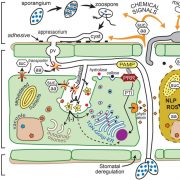
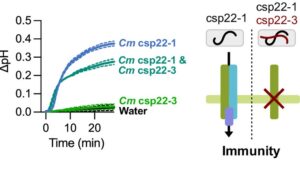 Host plants recognize diverse bacterial epitopes, known as microbe-associated molecular patterns (MAMPs), and respond with an immune reaction to control bacterial growth. However, most studies have focused on single bacterial epitopes, limiting our understanding of plant-bacteria interaction outcomes in real-world scenarios. Stevens et al. took an evolutionary approach to study five common MAMPs in 4,228 plant-associated bacteria. Their analysis showed that these MAMPs are evolutionarily constrained, despite variations in copy numbers (CNVs) and sequences, allowing for experimental testing of host immune responses. Among the MAMPs studied, cold shock proteins (CSPs) were highly diverse, both in CNVs and sequence divergence, and induced a range of immune responses in host plants, from non-immunogenic to highly immunogenic. Further experiments with a Clavibacter strain containing both immunogenic and non-immunogenic CSPs revealed that the non-immunogenic CSPs suppressed the activity of the immunogenic ones within the same strain. Additionally, cell lysates from this strain masked immune induction by other strains, including Streptomyces and the pathogenic Pseudomonas. This comprehensive resource on epitope variations and new insights into how epitopes interact to induce host immunity will help develop strategies to predictably manipulate host-pathogen interactions. (Summary by Arijit Mukherjee @ArijitM61745830) PNAS 10.1073/pnas.2319499121
Host plants recognize diverse bacterial epitopes, known as microbe-associated molecular patterns (MAMPs), and respond with an immune reaction to control bacterial growth. However, most studies have focused on single bacterial epitopes, limiting our understanding of plant-bacteria interaction outcomes in real-world scenarios. Stevens et al. took an evolutionary approach to study five common MAMPs in 4,228 plant-associated bacteria. Their analysis showed that these MAMPs are evolutionarily constrained, despite variations in copy numbers (CNVs) and sequences, allowing for experimental testing of host immune responses. Among the MAMPs studied, cold shock proteins (CSPs) were highly diverse, both in CNVs and sequence divergence, and induced a range of immune responses in host plants, from non-immunogenic to highly immunogenic. Further experiments with a Clavibacter strain containing both immunogenic and non-immunogenic CSPs revealed that the non-immunogenic CSPs suppressed the activity of the immunogenic ones within the same strain. Additionally, cell lysates from this strain masked immune induction by other strains, including Streptomyces and the pathogenic Pseudomonas. This comprehensive resource on epitope variations and new insights into how epitopes interact to induce host immunity will help develop strategies to predictably manipulate host-pathogen interactions. (Summary by Arijit Mukherjee @ArijitM61745830) PNAS 10.1073/pnas.2319499121