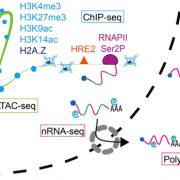
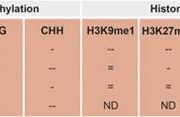
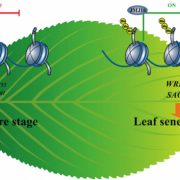
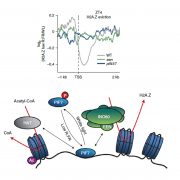
Non-CG methylation mechanism in Marchantia polymorpha ($) (Plant Cell Physiol.)
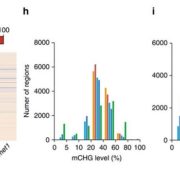
 Previous reports have shown that M. polymorpha DNA methylation status varies during its life cycle and it contains only one copy of the METHYLTRANSFERASE1 (MET1) orthologous gene. MET1 is responsible of CG methylation in angiosperms. Ikeda and collaborators generated MpMET mutants using the CRISPR/CAS9 system. The mutants showed abnormal shape, growth delay, reduced size and organs for asexual and sexual reproduction were not formed. Further analysis showed DNA hypomethylation of transposon regions. Bisulfite sequencing (BS-seq) was used to analyze wide genome methylation in the mutants; finding a 0,5% decreased methylation compared to wild type. Non-CG methylation was increased in transposon elements (TE) in mutants. Furthermore, CAG methylation was more efficient compared to CTG in the wild type, resulting on a asymmetrical methylation. On the other hand, the lack of CG methylation in the mutants resulted in a symmetrical methylation of CAG and CTG sites. (Summary by Cecilia Vasquez-Robinet). Plant & Cell Physiology 10.1093/pcp/pcy161
Previous reports have shown that M. polymorpha DNA methylation status varies during its life cycle and it contains only one copy of the METHYLTRANSFERASE1 (MET1) orthologous gene. MET1 is responsible of CG methylation in angiosperms. Ikeda and collaborators generated MpMET mutants using the CRISPR/CAS9 system. The mutants showed abnormal shape, growth delay, reduced size and organs for asexual and sexual reproduction were not formed. Further analysis showed DNA hypomethylation of transposon regions. Bisulfite sequencing (BS-seq) was used to analyze wide genome methylation in the mutants; finding a 0,5% decreased methylation compared to wild type. Non-CG methylation was increased in transposon elements (TE) in mutants. Furthermore, CAG methylation was more efficient compared to CTG in the wild type, resulting on a asymmetrical methylation. On the other hand, the lack of CG methylation in the mutants resulted in a symmetrical methylation of CAG and CTG sites. (Summary by Cecilia Vasquez-Robinet). Plant & Cell Physiology 10.1093/pcp/pcy161