Network paths for nitrogen response in Arabidopsis (Nature Comms)
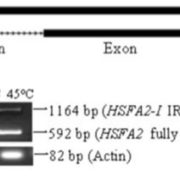
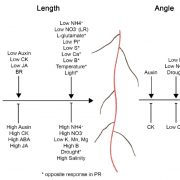
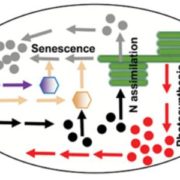
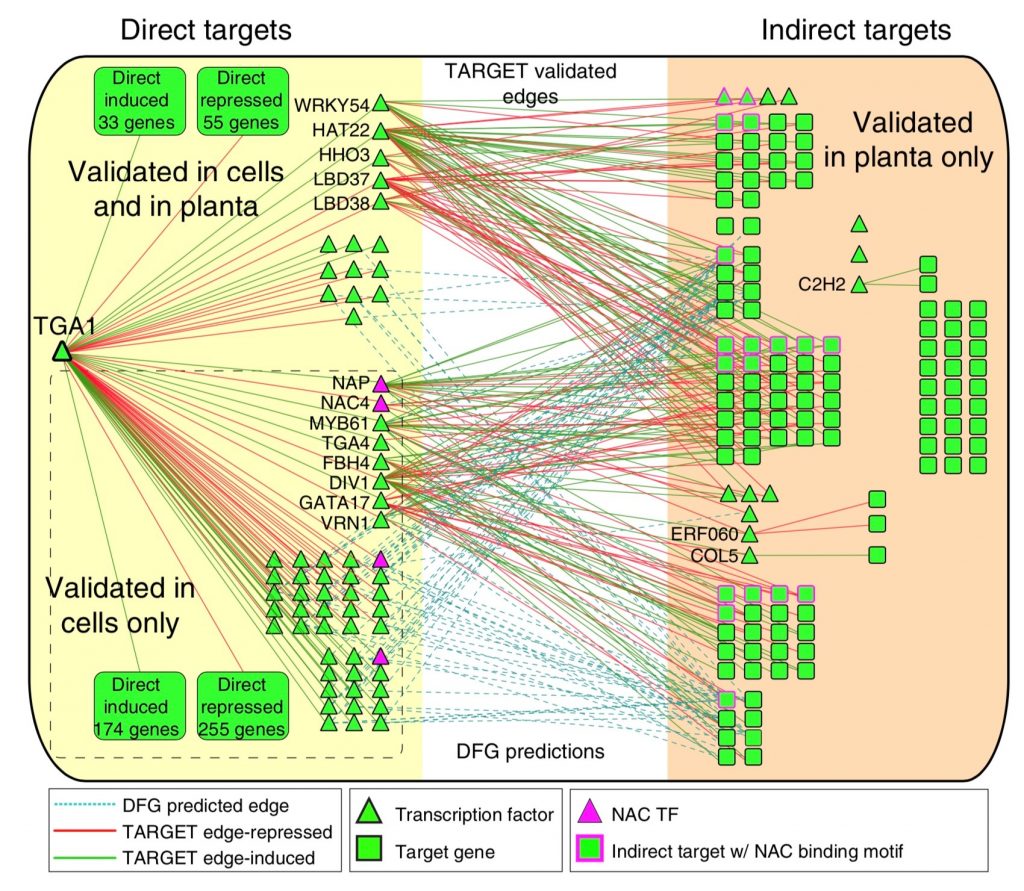 A goal of systems biology is to study how transcription factors participate in gene regulatory networks (GRN) underlying biological processes. In a recent report, Brooks et al. used network science to uncover how transcription factors mediate the early response to nitrogen (N). Authors identified the direct targets of 33 transcription factors using an in vivo approach called TARGET (Transient Assay Reporting Genome-wide Effects of Transcription factors), which detects TF-induced changes in gene expression. TARGET-data provides an excellent complementation to previously reported yeast one-hybrid (Y1H) and DNA affinity purification sequencing (DAP-seq) datasets. With all these tools together, a better understanding of TF-DNA binding can be achieved. Using Network Walking (a method for identifying the path of a TF to other downstream TFs and indirect targets), the authors reconstruct the order of TF-target interactions underlying the gene regulatory networks for TGA1 and CRF4, two important regulators of N response. There is still a lot of data to explore since 24 of the tested TF have an unknown role in N signaling. The generation of this network could have a direct impact on understanding and manipulation of GRN underlying nitrogen use in plants. (Summary by Humberto Herrera-Umbaldo) Nature Comms. 10.1038/s41467-019-09522-1
A goal of systems biology is to study how transcription factors participate in gene regulatory networks (GRN) underlying biological processes. In a recent report, Brooks et al. used network science to uncover how transcription factors mediate the early response to nitrogen (N). Authors identified the direct targets of 33 transcription factors using an in vivo approach called TARGET (Transient Assay Reporting Genome-wide Effects of Transcription factors), which detects TF-induced changes in gene expression. TARGET-data provides an excellent complementation to previously reported yeast one-hybrid (Y1H) and DNA affinity purification sequencing (DAP-seq) datasets. With all these tools together, a better understanding of TF-DNA binding can be achieved. Using Network Walking (a method for identifying the path of a TF to other downstream TFs and indirect targets), the authors reconstruct the order of TF-target interactions underlying the gene regulatory networks for TGA1 and CRF4, two important regulators of N response. There is still a lot of data to explore since 24 of the tested TF have an unknown role in N signaling. The generation of this network could have a direct impact on understanding and manipulation of GRN underlying nitrogen use in plants. (Summary by Humberto Herrera-Umbaldo) Nature Comms. 10.1038/s41467-019-09522-1