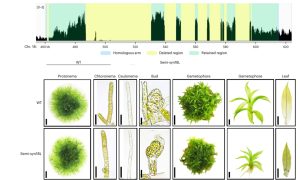
Moss phenotype unaffected by removal of repetitive sequences from genome
Genome complexity in multicellular organisms is often associated with repetitive sequences from Transposable Elements (TEs). TE function, importance for genome integrity, and dispensability have not been completely characterized. In prokaryotes and simple eukaryotes, genome synthesis (production of synthetic chromosomes and genomes) has proven to be a useful tool to understand genome organisation and function. As a prelude to the development of synthetic genomes in a multicellular organism. Physcomitrium (Physcomitrella) patens is a well-established multicellular model organism that is more complex than unicellular organisms due to greater genome size, gene number and epigenetic diversity. Chen and collaborators investigated the effect of decreased repetitive sequences by eliminating 55.8% of a 155 kb region of chromosome 18 in Physcomitrella. They used various methods to reassemble the DNA, including homologous recombination which was highly effective. Surprisingly, the phenotype of the transformed plant was comparable to the wild type (WT) even when growing under salt and osmotic stress. There was not much difference in histone marks related to active transcription between the synthetic and WT genomic regions, however marks associated with TEs abundance (H3K9me2) were almost completely removed, probably due to the lack of repetitive elements. On the other hand, chromatin interaction did show changes in the redesigned and neighbouring regions, demonstrating that genome redesign can alter chromosome conformation. These studies show that repetitive sequences can be removed without affecting morphology, development, and stress resistance, paving the way for the Synthetic Moss (SynMoss) project. (Summary by Rigel Salinas-Gamboa @Rigelitactica). Nature Plants 10.1038/s41477-023-01595-7