Modifying ripening through modular transcription
Sophia G. Zebell
Howard Hughes Medical Institute, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA
Manipulation of ripening, the process that turns hard, flavorless green tomatoes (Solanum lycopersicum) into the juicy, aromatic red fruits enjoyed worldwide, is of particular commercial and biological importance. Significant transcriptional reprogramming is required for synchronized cell wall softening and production of pigment, flavor, and aromatic compounds. A number of transcriptional regulators of ripening have been identified and studied extensively for their use in efforts to control ripening and extend the shelf life of fruits (Wang et al. 2020).
Due to its particularly useful nonripening phenotype, in which fruits fail to change color or soften, the tomato mutant ripening inhibitor (rin) has a long history of utilization in breeding and improvement programs. Mapped to the locus of a gene encoding a MADS-box transcription factor (TF), rin’s phenotype suggests loss of function of a key transcriptional activator of ripening, upstream of the ethylene burst thought to be required for all of the characteristic ripening phenotypes (Vrebalov et al. 2002). However, recent studies have clarified that in fact the RIN protein is dispensable for some ethylene production and the onset of ripening, as true null mutants begin to soften and turn orange with similar timing to wild-type plants (Ito et al. 2017). Instead, the complete inhibition of ripening observed in rin is due to a structural rearrangement that causes fusion of RIN with a nearby gene, leading to a novel repressor version of the protein (Ito et al. 2017, Li et al. 2018).
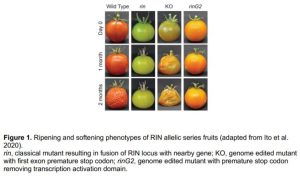
In this issue, Ito and colleagues present an allelic series of rin mutants, including a key C-terminal truncation allele obtained by CRISPR editing (Ito et al. 2020). By removing the transcription activation domain from the DNA-binding and protein–protein interaction domains of RIN, the truncated RIN alleles (rinG2), along with previously published null mutants, reveal a novel role for RIN as a repressor of tomato over-softening. Like those of rin, rinG2 fruits have an extended shelf life and reduced softening, whereas RIN knock out (KO) fruits soften much more quickly than wild type (Figure 1). Using transcriptomics, the authors characterize gene expression in wild-type, rin, rinG2, and KO plants, demonstrating that, unlike rin, knock out and truncated RIN alleles have normal induction of many ripening-associated genes, such as those responsible for carotenoid production. They are also able to leverage transcriptomics to demonstrate the potential for targeted editing and second site mutations to effectively harness the useful rinG2 traits, such as enhanced shelf stability, while mitigating undesirable pleiotropic effects, such as low lycopene accumulation in fruit from rinG2 plants.
Ito et al. propose a model in which rinG2 forms normal tetrameric complexes with its MADS-box TF partners but these complexes lack transcription activation activity, leading to reduced fruit softening and incomplete ripening. They support this model with in vitro protein–protein interaction data and ChIP of the promoter of a key fruit-softening regulator, where they find no binding of the complex in KO mutants but wild type-level complex binding in rinG2 plants. To reconcile the strong nonripening phenotype of rin, a key aspect of this model is redundancy between RIN and an unidentified ethylene-inducible transcription factor for the induction of most ripening-associated genes. These genes fail to be induced in rin and rinG2 mutants due to the dominant-negative presence of non-activating TF complexes at their promoters. Future studies will be necessary to identify the ethylene-inducible transcription regulator and the mechanism by which RIN acts as a repressor of over-softening.
The author’s model emphasizes partially overlapping roles for ethylene and RIN in fruit ripening regulation, further refining our understanding of ripening as a pleiotropic process with distinct modules of regulation and shared central transcription factors. The advances in this study were made possible by detailed analysis of an allelic series of mutations, highlighting the great potential of revisiting early plant genetic studies with CRISPR-Cas9 technology. In particular, the evolving model of RIN regulation of ripening emphasizes the need not only for true null mutations, but also for editing-based hypothesis testing to effectively interpret the function of proteins through classical reductionist genetics.
Ito, Yasuhiro, Ayako Nishizawa-Yokoi, Masaki Endo, Masafumi Mikami, Yoko Shima, Nobutaka Nakamura, Eiichi Kotake-Nara, Susumu Kawasaki, and Seiichi Toki. 2017. “Re-Evaluation of the Rin Mutation and the Role of RIN in the Induction of Tomato Ripening.” Nature Plants 3 (11): 866–74. https://doi.org/10.1038/s41477-017-0041-5.
Ito, Yasuhiro, Yasuyo Sekiyama, Hiroko Nakayama, Ayako Nishizawa-Yokoi, Masaki Endo, Yoko Shima, Nobutaka Nakamura, et al. 2020. “Allelic Mutations in the Ripening-Inhibitor (RIN) Locus Generate Extensive Variation in Tomato Ripening.” Plant Physiology, January. https://doi.org/10.1104/pp.20.00020.
Li, Shan, Huijinlan Xu, Zheng Ju, Dongyan Cao, Hongliang Zhu, Daqi Fu, Donald Grierson, Guozheng Qin, Yunbo Luo, and Benzhong Zhu. 2018. “The RIN-MC Fusion of MADS-Box Transcription Factors Has Transcriptional Activity and Modulates Expression of Many Ripening Genes.” Plant Physiology 176 (1): 891–909. https://doi.org/10.1104/pp.17.01449.
Vrebalov, Julia, Diane Ruezinsky, Veeraragavan Padmanabhan, Ruth White, Diana Medrano, Rachel Drake, Wolfgang Schuch, and Jim Giovannoni. 2002. “A MADS-Box Gene Necessary for Fruit Ripening at the Tomato Ripening-Inhibitor (Rin) Locus.” Science 296 (5566): 343–46. https://doi.org/10.1126/science.1068181.
Wang, Rufang, Gerco C. Angenent, Graham Seymour, and Ruud A. de Maagd. 2020. “Revisiting the Role of Master Regulators in Tomato Ripening.” Trends in Plant Science 25 (3): 291–301. https://doi.org/10.1016/j.tplants.2019.11.005.



