Modeling the metabolism of Arabidopsis thaliana: Application of network decomposition and network reduction in the context of Petri nets
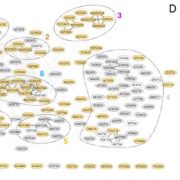
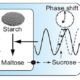
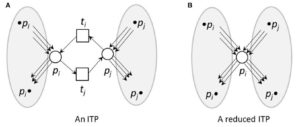 Front. Genetics. In this paper, the authors model the metabolism in Arabidopsis using Petri nets (PN). PNs are dynamic models that use “tokens” to represent movement between the places and edges of the model, representing the products of a reaction. The model was simplified using reduction techniques, and divided into four subnetworks: sucrose, citrate, UTP, and shikimate. The model provides an interpretation framework for the whole metabolic network and suggests some steady-state pathways and between-subnetwork dependencies. Front. Genetics 10.3389/fgene.2017.00085
Front. Genetics. In this paper, the authors model the metabolism in Arabidopsis using Petri nets (PN). PNs are dynamic models that use “tokens” to represent movement between the places and edges of the model, representing the products of a reaction. The model was simplified using reduction techniques, and divided into four subnetworks: sucrose, citrate, UTP, and shikimate. The model provides an interpretation framework for the whole metabolic network and suggests some steady-state pathways and between-subnetwork dependencies. Front. Genetics 10.3389/fgene.2017.00085