Measuring both microbial load and diversity with a single amplicon sequencing library (bioRxiv)
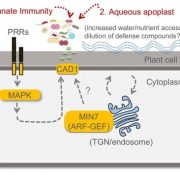
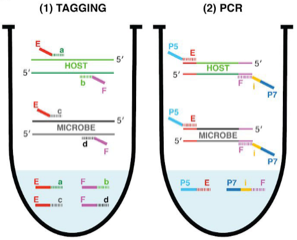 Amplicon sequencing of microbial DNA is a gold standard for analyzing the relative abundance of microbes in host-associated microbiomes. To gain more accurate insights into microbiome changes, it is crucial to know the absolute abundance of microbes, which can be analyzed by integrating relative abundance information with the total microbial load measured, which is measured by an additional experiment. However, such an approach is typically time consuming and noisy. To overcome this limitation, Lundberg et al. developed a simple and cost-effective strategy to measure both relative and absolute abundance of host-associated microbiomes from a single sequencing library. This approach, named host-associated microbe PCR (hamPCR), employs two steps of PCR reactions: the first limited-cycle PCR tags host and microbial target genes, then the second PCR amplifies all tagged products, followed by next-generation sequencing. PCR amplification with the common primers that target the tags can minimize amplification biases caused by differential primer efficiencies. The authors implemented a clever strategy to preadjust the host-to-microbe ratio in a sequencing library without losing information, which helps improve accuracy and cost efficiency in analyzing samples dominated by the host or microbial DNA. The method performed well for three amplicons (plants, bacteria, and oomycetes) and is applicable to a crop with a large genome and a non-plant host (nematode worms). In summary, hamPCR is a robust and easy-to-implement method that offers an accurate picture of the host-associated microbiome. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2020.05.19.103937
Amplicon sequencing of microbial DNA is a gold standard for analyzing the relative abundance of microbes in host-associated microbiomes. To gain more accurate insights into microbiome changes, it is crucial to know the absolute abundance of microbes, which can be analyzed by integrating relative abundance information with the total microbial load measured, which is measured by an additional experiment. However, such an approach is typically time consuming and noisy. To overcome this limitation, Lundberg et al. developed a simple and cost-effective strategy to measure both relative and absolute abundance of host-associated microbiomes from a single sequencing library. This approach, named host-associated microbe PCR (hamPCR), employs two steps of PCR reactions: the first limited-cycle PCR tags host and microbial target genes, then the second PCR amplifies all tagged products, followed by next-generation sequencing. PCR amplification with the common primers that target the tags can minimize amplification biases caused by differential primer efficiencies. The authors implemented a clever strategy to preadjust the host-to-microbe ratio in a sequencing library without losing information, which helps improve accuracy and cost efficiency in analyzing samples dominated by the host or microbial DNA. The method performed well for three amplicons (plants, bacteria, and oomycetes) and is applicable to a crop with a large genome and a non-plant host (nematode worms). In summary, hamPCR is a robust and easy-to-implement method that offers an accurate picture of the host-associated microbiome. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2020.05.19.103937