Master MYCs: MYC2, the jasmonate signaling ‘master switch’
To optimize their fitness in the field, plants need to respond rapidly, specifically and dynamically to an ever-changing and often hostile environment. By integrating external environmental cues with endogenous developmental programs, phytohormones play a critical role in the cross-talk between signal transduction networks. Juxtaposed between the plant defense and development pathways is the oxylipin signaling molecule jasmonic acid and its derivatives, collectively known as jasmonates (JAs).
The JA signaling pathway is well studied in Arabidopsis thaliana. At its core are the JA receptor CORONATINE INSENSITIVE 1 (COI1); the basic helix-loop-helix (bHLH) transcription factor (TF) MYC2, which acts additively with its close homologs MYC3 and MYC4 as major regulators of diverse JA responses; and a group of jasmonate-ZIM domain (JAZ) proteins, which function as transcriptional repressors of MYC2/MYC3/MYC4. To induce JA-responsive genes, MYC2 forms a transcriptional activation complex with MED25, a subunit of the Mediator complex. Crucially, MED25 also interacts with COI1, thus bridging the JA receptor to MYC2-targetted promoters (An et al., 2017).
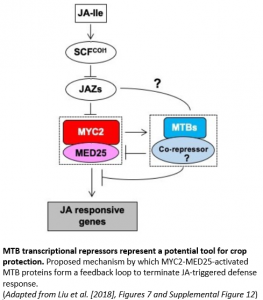
Given that sustained activation of JA signaling can severely inhibit plant growth, turning off JA signaling is as important as turning it on. In a new study, Li and colleagues (2018) describe an additional and unexpected function of MYC2 in forming an autoregulatory negative feedback circuit that regulates the termination of JA signaling in tomato (Solanum lycopersicum).
The first clue that MYC2 itself may be involved in the cessation of JA signaling came from their functional analysis of tomato plants overexpressing MYC2 (MYC2-OE). Given the established role of MYC2 in activating JA-responsive gene expression, the authors initially hypothesized that the MYC2-OE mutant would show enhanced JA-defense responses. Surprisingly, however, they observed that both wound-induced JA accumulation and expression levels of known defense targets of MYC2 were reduced in the MYC2-OE line.
Utilizing their existing ChIP-Seq dataset of the genome-wide binding sites of MYC2 (Du et al., 2017), the authors set out to discover possible downstream TF targets of MYC2 responsible for the observed negative regulation of JA signaling. They identified three MYC2-TARGETED BHLH TFs (MTB1, MTB2 and MTB3), homologs of the Arabidopsis JASMONATE-ASSOCIATED MYC2-LIKE (JAM) transcriptional repressors. Detailed functional characterization of MTB-RNAi and MTB1-OE plants demonstrated that MTB1–3 negatively regulate diverse aspects of known JA responses in tomato, including the wound response, anthocyanin accumulation, and defense responses to both herbivorous insects and necrotrophic pathogens. Indeed, the authors later showed that CRISPR/Cas9 mtb tomato lines have increased resistance to herbivore attack but with no visible alterations in other agronomical traits.
 Sequence analysis of the MTB proteins revealed that they share high similarity to MYC2 itself, which extends to both their DNA-binding and JAZ-interaction domains. Notably, MTB1 is capable of forming heterodimers with MYC2 in vitro. This finding hinted at a possible mechanism of action for these transcriptional repressors: disruption of the interaction of MYC2 with its co-activator MED25. The authors confirmed this using a combination of competitive pull-down assays showing that MTB1 does indeed interfere with MYC2–MED25 interaction, and ChIP-qPCR wherein MTB-RNAi and OE lines had altered enrichment of MED25 on the promoters of known targets. In fact, electrophoretic mobility shift assays revealed that MTB proteins possess a second molecular mechanism that impinges on the activity of the MYC2–MED25 transcriptional activation complex: MTB1 directly and competitively binds to the same G-box promoter regions as MYC2. Given that MTBs are capable of interacting with JAZ proteins, the authors speculate that a third mechanism involving repressor-repressor interactions may additionally be at play.
Sequence analysis of the MTB proteins revealed that they share high similarity to MYC2 itself, which extends to both their DNA-binding and JAZ-interaction domains. Notably, MTB1 is capable of forming heterodimers with MYC2 in vitro. This finding hinted at a possible mechanism of action for these transcriptional repressors: disruption of the interaction of MYC2 with its co-activator MED25. The authors confirmed this using a combination of competitive pull-down assays showing that MTB1 does indeed interfere with MYC2–MED25 interaction, and ChIP-qPCR wherein MTB-RNAi and OE lines had altered enrichment of MED25 on the promoters of known targets. In fact, electrophoretic mobility shift assays revealed that MTB proteins possess a second molecular mechanism that impinges on the activity of the MYC2–MED25 transcriptional activation complex: MTB1 directly and competitively binds to the same G-box promoter regions as MYC2. Given that MTBs are capable of interacting with JAZ proteins, the authors speculate that a third mechanism involving repressor-repressor interactions may additionally be at play.
Crucially, the formation of this MYC2–MTB autoregulatory feedback loop is tightly controlled with wound-induced expression of the MTB genes temporally delayed relative to MYC2 (see Figure). This delay neatly ensures that the ‘off-switch’ for terminating JA-mediated responses is pre-programmed into the induction phase (‘on-switch’) of JA signaling whilst still allowing an appropriate JA response to proceed. Such negative autoregulatory feedback loops are frequently found in inductive pathways, functioning as a trip-switch to shut down the deleterious effects of a sustained response.
The work of Li and colleagues has potential applications in sustainable crop production since it may offer a strategy to engineer the relationships between growth and defense.
REFERENCES
An, C., Li, L., Zhai, Q., You, Y., Deng, L., Wu, F., Chen, R., Jiang, H., Wang, H., Chen, Q., and Li, C. (2017). Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proceedings of the National Academy of Sciences 114: E8930–E8939.
Du, M., Zhao, J., Tzeng, D.T.W., Liu,Y., Deng, L., Yang, T., Zhai, Q., Wu, F., Huang, Z., Zhou, M., Wang, Q., Chen, Q., Zhong, S., Li, C-B., Li C. (2017). MYC2 Orchestrates a Hierarchical Transcriptional Cascade That Regulates Jasmonate-Mediated Plant Immunity in Tomato. Plant Cell 29: 1883–1906.
Liu, Y., Du, M., Deng, L., Shen, J., Fang, M., Chen, Q., Lu, Y., Wang, Q., Li, C., and Zhai, Q. (2019). MYC2 Regulates the Termination of Jasmonate Signaling via an Autoregulatory Negative Feedback Loop. Plant Cell; doi: 10.1105/tpc.18.00405