MapMan4: A refined protein classification and annotation framework applicable to multi-omics data analysis (Mol Plant)
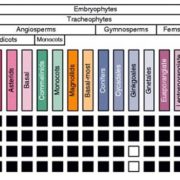
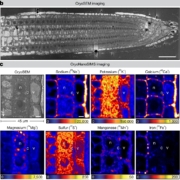
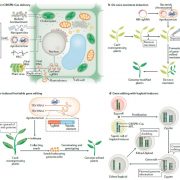
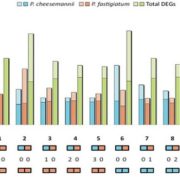
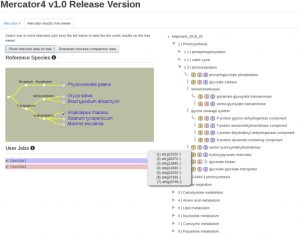 Revolutions in omics technologies have rapidly increased the number of plant genomes, transcriptomes and proteomes available. However, protein sequences need to be assigned a biological function to provide useful insights for comparative genomics and transcriptomics. Here, Schwacke et al present MapMan4, a tool to classify proteins and provide ontology information for omics data as well as an associated package, Mercator, an online protein annotation tool. MapMan4 is compared against other ontology tools including Gene Ontology (GO) and KEGG. Arabidopsis thaliana is annotated at a success rate of 47% using Mercator compared to 32% using a KEGG annotation. The authors demonstrate these techniques by studying gene loss in parasitic plants. More information about MapMan4 and Mercator is available (Summary by Alex Bowles) Molecular Plant 10.1016/j.molp.2019.01.003
Revolutions in omics technologies have rapidly increased the number of plant genomes, transcriptomes and proteomes available. However, protein sequences need to be assigned a biological function to provide useful insights for comparative genomics and transcriptomics. Here, Schwacke et al present MapMan4, a tool to classify proteins and provide ontology information for omics data as well as an associated package, Mercator, an online protein annotation tool. MapMan4 is compared against other ontology tools including Gene Ontology (GO) and KEGG. Arabidopsis thaliana is annotated at a success rate of 47% using Mercator compared to 32% using a KEGG annotation. The authors demonstrate these techniques by studying gene loss in parasitic plants. More information about MapMan4 and Mercator is available (Summary by Alex Bowles) Molecular Plant 10.1016/j.molp.2019.01.003