Many plant transcription factor families have evolutionarily conserved binding motifs
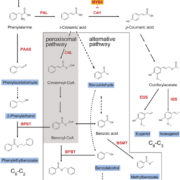
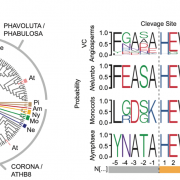
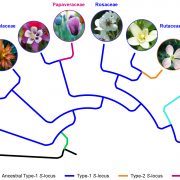
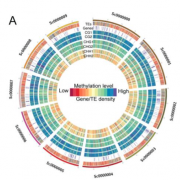
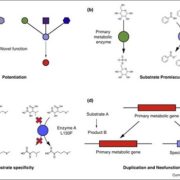
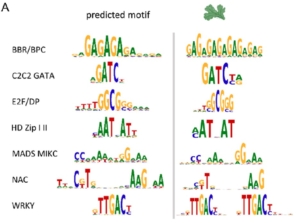 The regulated expression of genes is fundamental to all biological processes, including development, cell growth, and responses to environmental signals. Transcription factors (TFs) are sequence-specific DNA-binding proteins that play a central role in transcriptional regulation by directly interacting with cis-regulatory elements of gene targets. During evolution, the specificity of these interactions can shift due to genome variations or gene duplications. Such changes have been particularly pronounced in yeast, whereas they appear more conserved in flies and humans. To explore this in plants, Zenker and colleagues revisited existing plant TF databases and recalculated TF binding consensus sequences using a standardized pipeline. By comparing angiosperm species with the bryophyte Marchantia polymorpha, their analyses revealed a spectrum of sequence conservation across different TF families, with some binding motifs showing remarkable conservation, tracing back 450 million years. While the relative contributions of cis– and trans-regulation might be challenging to disentangle, this comprehensive catalog of plant TF binding motifs offers valuable insights for studying neofunctionalization and improving TF annotation in emerging plant models. (Summary by Ching Chan @ntnuchanlab) bioRxiv 10.1101/2024.10.31.621407
The regulated expression of genes is fundamental to all biological processes, including development, cell growth, and responses to environmental signals. Transcription factors (TFs) are sequence-specific DNA-binding proteins that play a central role in transcriptional regulation by directly interacting with cis-regulatory elements of gene targets. During evolution, the specificity of these interactions can shift due to genome variations or gene duplications. Such changes have been particularly pronounced in yeast, whereas they appear more conserved in flies and humans. To explore this in plants, Zenker and colleagues revisited existing plant TF databases and recalculated TF binding consensus sequences using a standardized pipeline. By comparing angiosperm species with the bryophyte Marchantia polymorpha, their analyses revealed a spectrum of sequence conservation across different TF families, with some binding motifs showing remarkable conservation, tracing back 450 million years. While the relative contributions of cis– and trans-regulation might be challenging to disentangle, this comprehensive catalog of plant TF binding motifs offers valuable insights for studying neofunctionalization and improving TF annotation in emerging plant models. (Summary by Ching Chan @ntnuchanlab) bioRxiv 10.1101/2024.10.31.621407