Major impacts of widespread structural variation on gene expression and crop improvement in tomato (Cell)
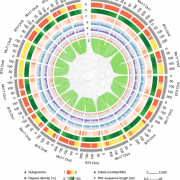
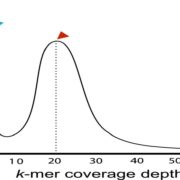
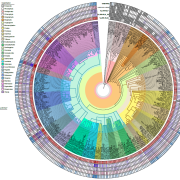
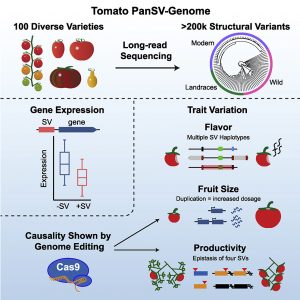 Structural Variants (SVs) are large genomic deletions, insertions, and duplications with underexplored roles in determining plant phenotypes. Recognizing the extent to which SVs define quantitative trait variation was previously constrained by the fact that popular short-read sequencing technologies preferentially characterize smaller variants. However, innovative long-read technologies are facilitating extensive population-level surveys of SVs. Here, Alonge, Wang et al. present panSV, a tomato SV pangenome formed from long-read sequencing of 100 accessions that span phylogenetic diversity. Remarkably, over 238,000 SVs were found, with many shaped by transposons. Across the 100 genomes, 50% of SVs overlap with genes/regulatory sequences, and 95% of genes possess an SV within 5kbp of coding sequences. By employing panSV, 14 new reference genomes, RNA-seq, and CRISPR-Cas9, the authors functionally linked SVs to three major domestication and breeding traits: an unpalatable smoky flavor, a determinant of fruit weight, and jointless fruit pedicels. This study represents the release of the most comprehensive crop SV pangenome, and the abundant resources generated will expedite acknowledgment of the significance of SVs in evolution, domestication, and quantitative trait variation, undoubtedly advancing future crop improvement efforts. (Summary by Caroline Dowling @CarolineD0wling) Cell 10.1016/j.cell.2020.05.021
Structural Variants (SVs) are large genomic deletions, insertions, and duplications with underexplored roles in determining plant phenotypes. Recognizing the extent to which SVs define quantitative trait variation was previously constrained by the fact that popular short-read sequencing technologies preferentially characterize smaller variants. However, innovative long-read technologies are facilitating extensive population-level surveys of SVs. Here, Alonge, Wang et al. present panSV, a tomato SV pangenome formed from long-read sequencing of 100 accessions that span phylogenetic diversity. Remarkably, over 238,000 SVs were found, with many shaped by transposons. Across the 100 genomes, 50% of SVs overlap with genes/regulatory sequences, and 95% of genes possess an SV within 5kbp of coding sequences. By employing panSV, 14 new reference genomes, RNA-seq, and CRISPR-Cas9, the authors functionally linked SVs to three major domestication and breeding traits: an unpalatable smoky flavor, a determinant of fruit weight, and jointless fruit pedicels. This study represents the release of the most comprehensive crop SV pangenome, and the abundant resources generated will expedite acknowledgment of the significance of SVs in evolution, domestication, and quantitative trait variation, undoubtedly advancing future crop improvement efforts. (Summary by Caroline Dowling @CarolineD0wling) Cell 10.1016/j.cell.2020.05.021