It was a Great, Green Year: Identification of a Chlorophyll Dephytylase That Functions in Chlorophyll Turnover
IN BRIEF by Jennifer Mach [email protected]
Green may have been the Pantone Color of the Year for 2013 (http://www.pantone.com/color-of-the-year-2013), but 2016 was a great year for papers on chlorophyll research, at The Plant Cell and beyond. In this year, we saw a pile of interesting papers examining chlorophyll degradation, including its regulatory mechanisms. For example, to identify transcription factors upstream of the key enzyme pheophorbide a oxygenase (PAO), which is a rate-limiting factor in chlorophyll breakdown, Ghandchi and co-authors (2016) used regression modeling and correlation analysis to look for factors that co-expressed with PAO under a variety of environmental conditions. They identified a correlation with specific transcription factors and a regulatory network that integrates stimuli from abiotic stresses (such as freezing and drought) and senescence to modulate specific transcription factors that control expression of PAO and other genes, and thus regulate chlorophyll breakdown. 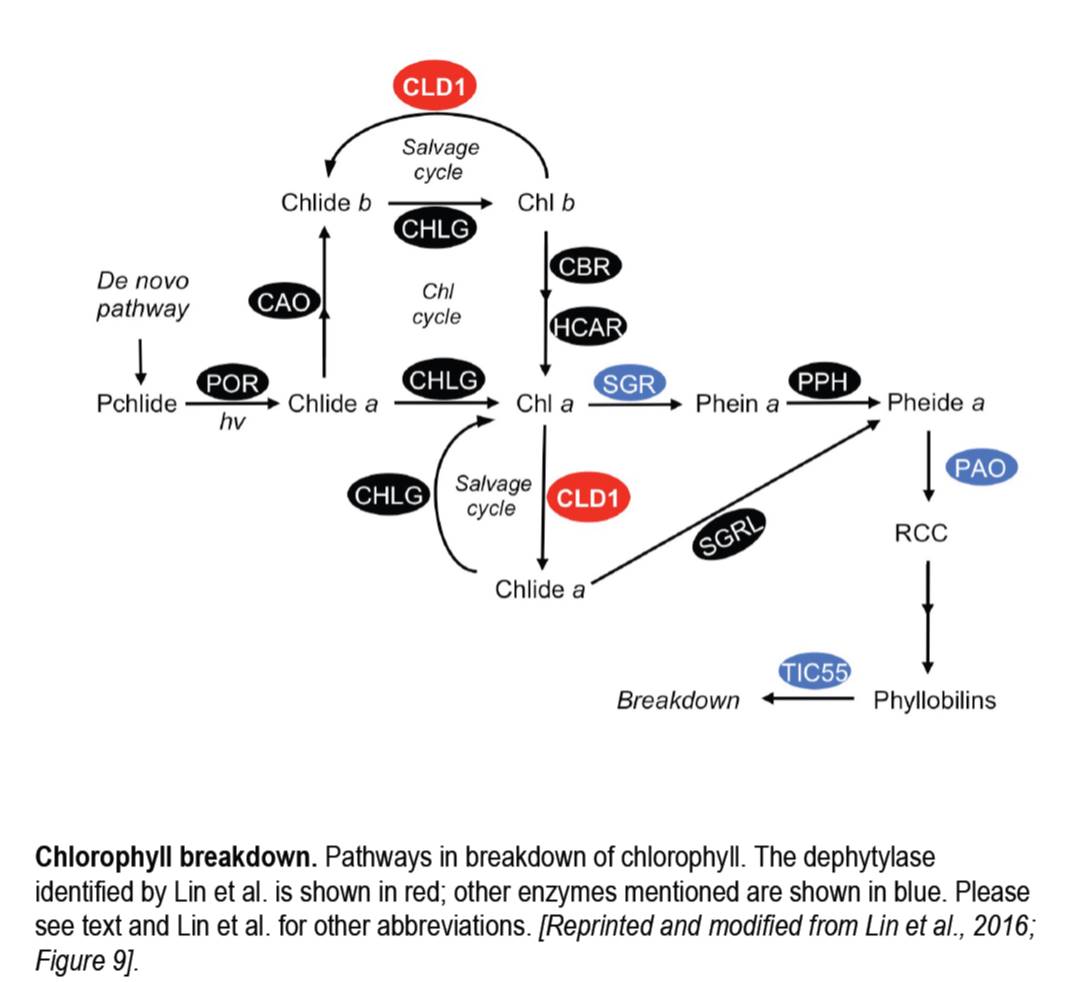
In another paper that generated some discussion, Wu and co-authors (2016) report that NON-YELLOWING2/STAY-GREEN2 (NYE2/SGR2), a close homolog of SGR/NYE1 (Mendel’s green cotyledon gene) functions as a positive regulator of chlorophyll breakdown and present several observations that are inconsistent with results from Sakuraba et al. (2014), specifically regarding the phenotypes of nye1 nye2 double mutants, which Wu et al. found to have a strong staygreen phenotype. These discrepancies, possibly due to growth conditions or genetic effects, and the role of NYE2/SGR2 await further clarification.
Enzymes also received a bit of attention in 2016, with the discovery of several missing links in the chlorophyll breakdown pathway (figure). For example, Shimoda et al. (2016) revealed that STAY-GREEN (SGR) functions as a magnesium dechelatase, which converts chlorophyll a to pheophytin a and also functions in degradation of the photosystems. Further along the chlorophyll breakdown pathway, Hauenstein and co-authors found an unexpected role for the chloroplast protein TRANSLOCON AT THE INNER CHLOROPLAST ENVELOPE 55 (TIC55) in the degradation of phyllobilins. TIC55 had been implicated in redox-regulated protein import in the chloroplast, making this finding surprising.
To cap off this year, in the December issue of The Plant Cell, Lin et al., (2016) report the exciting, somewhat serendipitous discovery of another missing enzyme, the activity that removes chlorophyll’s phytol tail during chlorophyll turnover. The authors started their research by examining a semidominant mutant that produces a heatsensitive phenotype and ended up identifying CHLOROPHYLL DEPHYTYLASE1 (CLD1). After cloning the locus responsible for the mutant phenotype, the authors speculated that the encoded enzyme might act on chlorophyll, based on its conservation and similarity to pheophytinase (PPH, see figure), particularly in the active site region. In vitro assays showed that CLD1 can dephytylate chlorophyll a and b, and pheophytin a, and that the mutant protein had higher activity than wild-type CLD1. Excess CLD1 activity caused mutant plants to accumulate chlorophyllides after heat shock and suppression of CLD1 by an artificial microRNA also caused plants to be sensitive to heat stress, with damage to Photosystem II, indicating the importance of chlorophyll turnover in photosystem stability.
Pantone might choose puce or magenta as the color of the year for 2017, but at The Plant Cell, we choose green every year.
Here’s to another great, green year!
REFERENCES
Ghandchi, F.P., Caetano-Anolles, G., Clough, S.J., and Ort, D.R. (2016). Investigating the Control of Chlorophyll Degradation by Genomic Correlation Mining. PLOS ONE 11(9): e0162327.
Hauenstein, M., Christ, B., Das, A., Aubry, S., and Hörtensteiner, S. (2016). A role for TIC55 as a hydroxylase of phyllobilins, the products of chlorophyll breakdown during plant senescence. Plant Cell 10.1105/tpc.16.00630.
Lin, Y.-P., Wu, M.-C., and Charng, Y.-y. (2016). Identification of Chlorophyll Dephytylase Involved in Chlorophyll Turnover in Arabidopsis. Plant Cell 10.1105/tpc.16.00478.
Sakuraba, Y., Park, S.Y., Kim, Y.S., Wang, S.H., Yoo, S.C., Hortensteiner, S., and Paek, N.C. (2014). Arabidopsis STAYGREEN2 is a negative regulator of chlorophyll degradation during leaf senescence. Mol. Plant 7:1288–1302.
Shimoda, Y., Ito, H., and Tanaka, A. (2016). Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesiumdechelatase. Plant Cell 28: 2147-2160.
Wu, S., Li, Z., Yang, L., Xie, Z., Chen, J., Zhang, W., Liu, T., Gao, S., Gao, J., Zhu, Y., Xin, J., Ren, G., and Benke Kuai, B. (2016) NON-YELLOWING2 (NYE2), a Close Paralog of NYE1, Plays a Positive Role in Chlorophyll Degradation in Arabidopsis. Molecular Plant 9: 624 – 627.




Leave a Reply
Want to join the discussion?Feel free to contribute!