In the Pale Red Light: Control of pre-mRNA Splicing by RRC1 and SFPS
Plants have armed themselves with a battalion of photoreceptors to cope with changes in light quantity, quality and direction. Light perception is especially critical when the seedling first emerges from the darkness of the soil and engages the red/far-red light photoreceptors phytochromes (phys). An avalanche of genetic screens has fleshed out phy signaling cascade over the past three decades, and has exposed control at multiple levels: transcription, translation, mRNA half-life, and most recently pre-mRNA splicing (Shikata et al., 2012).
This raises a fundamental question: how do phys achieve signal specificity? Perhaps they do so, in part, by combining multiple factors to generate the desired outcome. In their article, Xin et al. (2019) shed some light on one such factor: the RNA-binding protein and splicing factor RRC1 (for REDUCED RED LIGHT RESPONSES IN cry1cry2 BACKGROUND 1).
In a previous study, the authors had identified another factor, SFPS (SPLICING FACTOR FOR PHYTOCHROME SIGNALING; Xin et al., 2017). sfps mutants were impaired in a number of red-light mediated responses, including control of hypocotyl elongation, apical hook opening and flowering time. The identity of SFPS came as a surprise: the gene encoded the plant orthologue of human splicing factor SPF45. Transcriptome sequencing revealed that SFPS influenced the light-mediated alternative splicing of hundreds of transcripts (Xin et al., 2017).
In the current study, the authors use affinity purification followed by mass spectrometry to identify SFPS-interacting proteins, and focus here on RRC1, another splicing factor. Their light-independent physical interaction is confirmed by yeast two-hybrid, in vitro GST pull-downs and in vivo pull-downs of tagged proteins. Both proteins completely co-localize in nuclear speckles with each other and with U2 snRNP-associated components and partially with phyB in a light-dependent manner.
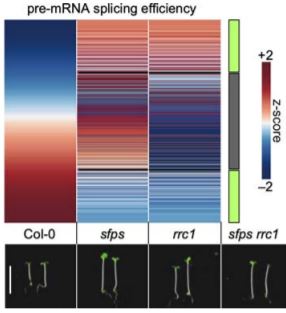
 SFPS and RCC1 likely work in the same pathway, as sfps and rrc1 single mutants and the sfps rrc1 double mutant have identical hypocotyl phenotypes in red light (see Figure). Although the rrc1 allele used here is hypomorphic, a stronger allele only produces 4% homozygous rrc1 plants from a heterozygous parent and has a comparable hypocotyl phenotype (Shikata et al., 2012).
SFPS and RCC1 likely work in the same pathway, as sfps and rrc1 single mutants and the sfps rrc1 double mutant have identical hypocotyl phenotypes in red light (see Figure). Although the rrc1 allele used here is hypomorphic, a stronger allele only produces 4% homozygous rrc1 plants from a heterozygous parent and has a comparable hypocotyl phenotype (Shikata et al., 2012).
Since RRC1 and SFPS are pre-mRNA splicing factors, the authors next explore the transcriptome and alternative splicing landscapes of the mutants in etiolated seedlings exposed (or not) to red light. They discover that either loss of function changes the expression of thousands of genes both in etiolated and red light-irradiated seedlings. The splicing pattern of thousands of genes is also disrupted in the rrc1 mutant, but only 20% of those genes are both differentially expressed and differentially spliced, independently of the light conditions. In fact, alternative splicing of pre-mRNAs in rrc1 is largely blind to the light status, in contrast to wild type, but very evocative of sfps (Xin et al., 2017). Despite their similar phenotypes, many more genes encounter defects in alternative splicing in rrc1 than in sfps, but the genes affected in both mutants largely experience the same change in splicing efficiency (see Figure).
To complicate things, the authors demonstrate that RRC1 expression and splicing are targets of SFPS, as were other splicing factors (Shikata et al., 2012; Xin et al., 2017), blurring the lines between direct and indirect effects. The emerging model suggests that phyB modulates the activity of RRC1 and SFPS (and additional factors), in turn targeting the splicing of many genes to regulate light responses.
References:
Shikata H, Shibata M, Ushijima T, Nakashima M, Kong SG, Matsuoka K, Lin C, Matsushita T (2012). The RS domain of Arabidopsis splicing factor RRC1 is required for phytochrome B signal transduction. Plant J. 70: 727-738.
Xin R, Kathare PK and Huq E (2019). Coordinated Regulation of Pre-mRNA Splicing by the SFPS-RRC1 Complex to Promote Photomorphogenesis. The Plant Cell. Published June 2019. DOI: https://doi.org/10.1105/tpc.18.00786
Xin R, Zhu L, Salomé PA, Mancini E, Marshall CM, Harmon FG, Yanovsky MJ, Weigel D, and Huq E (2017). SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis. PNAS 114: E7018-E7027.
Figure Legend.
The splicing factors SPFS and RRC1 both control hypocotyl elongation in red light. Their loss of function causes a deregulation in light-mediated pre-mRNA splicing efficiency of alternatively-spliced genes in similar (indicated by the green box on the right side of the heatmap) and opposite (gray box) direction. Differential splicing efficiency is shown as the z-score of the difference between splicing efficiency in the dark versus efficiency in red light [Adapted from Figure 5, Xin et al., 2019].